Retrovirus
Retrovirus (W)
A retrovirus is a type of virus that inserts a copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. Once inside the host cell's cytoplasm, the virus uses its own reverse transcriptase enzyme to produce DNA from its RNA genome, the reverse of the usual pattern, thus retro (backwards). The new DNA is then incorporated into the host cell genome by an integrase enzyme, at which point the retroviral DNA is referred to as a provirus. The host cell then treats the viral DNA as part of its own genome, transcribing and translating the viral genes along with the cell's own genes, producing the proteins required to assemble new copies of the virus.
Although retroviruses have different subfamilies, they have three basic groups. The oncoretroviruses (oncogenic retroviruses), the lentiviruses (slow retroviruses) and the spumaviruses (foamy viruses). The oncoretroviruses are able to cause cancer in some species, the lentiviruses able to cause severe immunodeficiency and death in humans and other animals, and the spumaviruses are benign and not linked to any disease in humans or animals.
Many retroviruses cause serious diseases in humans, other mammals, and birds. Human retroviruses include HIV-1 and HIV-2, the cause of the disease AIDS. Also, human T-lymphotropic virus (HTLV) causes disease in humans. The murine leukemia viruses (MLVs) cause cancer in mouse hosts. Retroviruses are valuable research tools in molecular biology, and they have been used successfully in gene delivery systems. |
| |
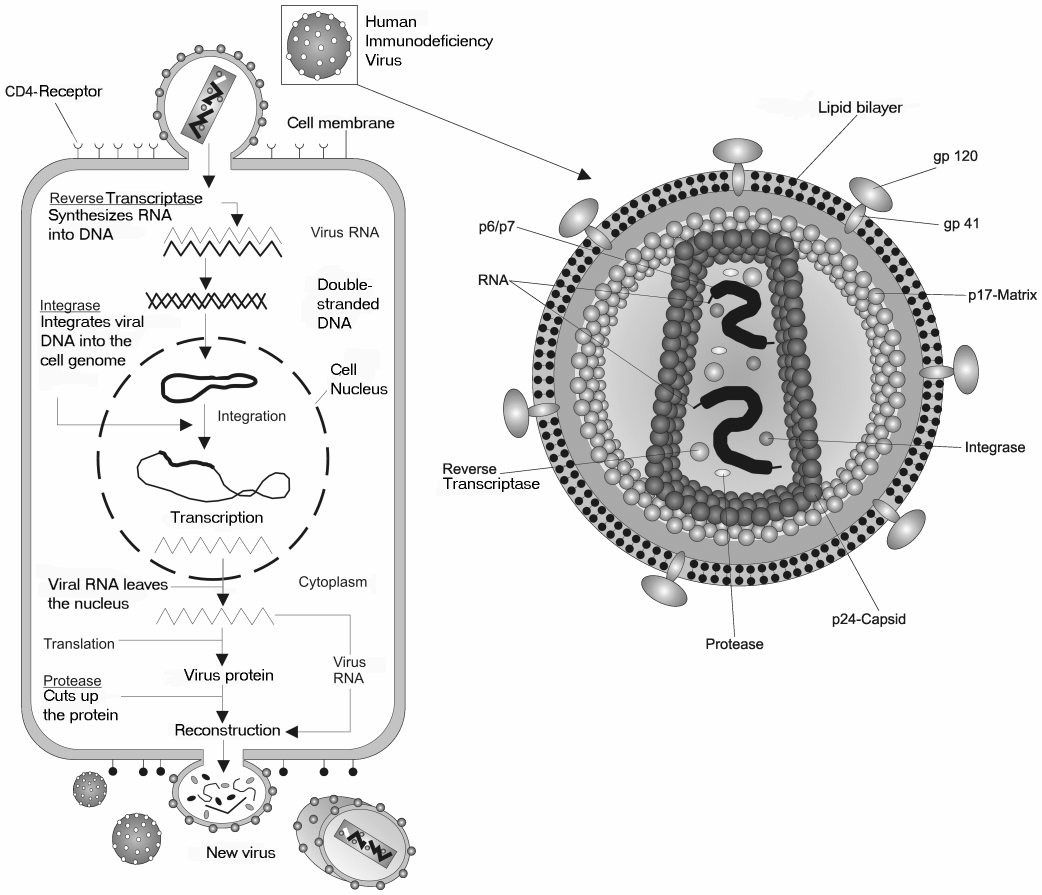
HIV retrovirus schematic of cell infection, virus production and virus structure. |
| |
|
|
|
| |
| |
Structure
|
Structure
Structure (W)
Virions of retroviruses consist of enveloped particles about 100 nm in diameter. The outer lipid envelope consists of glycoprotein. The virions also contain two identical single-stranded RNA molecules 7–10 kilobases in length.The two molecules are present as a dimer, formed by base pairing between complementary sequences. Interaction sites between the two RNA molecules have been identified as a "kissing -loop complex". Although virions of different retroviruses do not have the same morphology or biology, all the virion components are very similar.
The main virion components are:
- Envelope: composed of lipids (obtained from the host plasma membrane during the budding process) as well as glycoprotein encoded by the env gene. The retroviral envelope serves three distinct functions: protection from the extracellular environment via the lipid bilayer, enabling the retrovirus to enter/exit host cells through endosomal membrane trafficking, and the ability to directly enter cells by fusing with their membranes.
- RNA: consists of a dimer RNA. It has a cap at the 5' end and a poly(A) tail at the 3' end. Genomic RNA (gRNA) is produced as a result of host RNA polymerase II (Pol II) activity and by adding a 5' methyl cap and a 3' poly-A tail is processed as a host mRNA. The RNA genome also has terminal noncoding regions, which are important in replication, and internal regions that encode virion proteins for gene expression. The 5' end includes four regions, which are R, U5, PBS, and L. The R region is a short repeated sequence at each end of the genome used during the reverse transcription to ensure correct end-to-end transfer in the growing chain. U5, on the other hand, is a short unique sequence between R and PBS. PBS (primer binding site) consists of 18 bases complementary to 3' end of tRNA primer. L region is an untranslated leader region that gives the signal for packaging of the genome RNA. The 3' end includes 3 regions, which are PPT (polypurine tract), U3, and R. The PPT is a primer for plus-strand DNA synthesis during reverse transcription. U3 is a sequence between PPT and R, which serves as a signal that the provirus can use in transcription. R is the terminal repeated sequence at 3' end.
- Proteins: consisting of gag proteins, protease (PR), pol proteins, and env proteins.
- Group-specific antigen (gag) proteins are major components of the viral capsid, which are about 2000–4000 copies per virion. Gag possesses two nucleic acid binding domains, including matrix (MA) and nucleocapsid (NC). Specifically recognizing, binding, and packaging the retroviral genomic RNA into assembling virions is one of the important functions of Gag protein. Gag interactions with cellular RNAs also regulate aspects of assembly.The expression of gag alone gives rise to assembly of immature virus-like particles that bud from the plasma membrane. In all retroviruses the Gag protein is the precursor to the internal structural protein.
- Protease (pro) is expressed differently in different viruses. It functions in proteolytic cleavages during virion maturation to make mature gag and pol proteins. Retroviral Gag proteins are responsible for coordinating many aspects of virion assembly.
- Pol proteins are responsible for synthesis of viral DNA and integration into host DNA after infection.
- Env proteins play a role in association and entry of virions into the host cell. Possessing a functional copy of an env gene is what makes retroviruses distinct from retroelements. The ability of the retrovirus to bind to its target host cell using specific cell-surface receptors is given by the surface component (SU) of the Env protein, while the ability of the retrovirus to enter the cell via membrane fusion is imparted by the membrane-anchored trans-membrane component (TM). Thus it is the Env protein that enables the retrovirus to be infectious.
- Several protein species are associated with the RNA in the retrovirus virion. Nucleocapsid (NC) protein is the most abundant protein, which coats the RNA; while other proteins, present in much smaller amounts and have enzyme activities. Some enzyme activities that are present in the retrovirus virion includes RNA-dependent DNA polymerase (reverse transcriptase; RT), DNA-dependent DNA polymerase, Ribonuclease H (RNase H) Integrase and Protease. The retroviral RNases H encoded by all retroviruses, including HIV have been demonstrated to show three different modes of cleavage: internal, DNA 3′ end-directed, and RNA 5′ end-directed. All three modes of cleavage constitute roles in reverse transcription. Therefore, The RNase H activity is essential in several aspects of reverse transcription. The use of an RNase H activity during retroviral replication displays a unique strategy to copy a single-stranded RNA genome into a double-stranded DNA, since the minus-strand DNA are complementary and make base pairing to retrovirus genome in the first cycle of DNA synthesis. The RNase H ribonuclease activity is also required in the retroviral life cycle, since it generates and removes primers essential by the Reverse Transcriptase (RT) for the initiation of DNA synthesis. Retroviruses that are lacking RNase H activity are noninfectious.
|

|
|
|
|
Genomic structure
Genomic structure (W)
The retroviral genome is packaged as viral particles. These viral particles are dimers of single-stranded, positive-sense, linear RNA molecules.
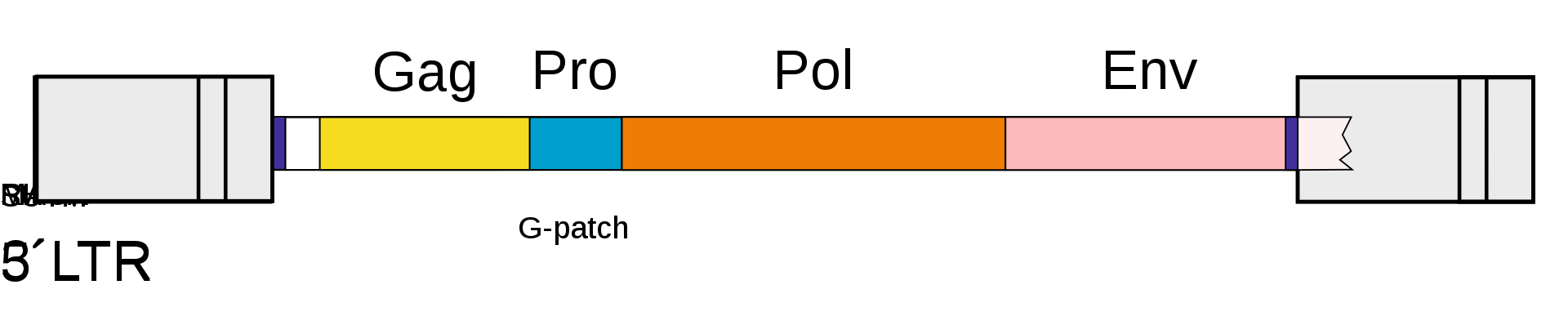
The genomic and subgenomic organization of a prototypical retrovirus. Abbreviations are explained in the file description. |
Retroviruses (and orterviruses in general) follow a layout of 5'–gag–pro–pol–env–3' in the RNA genome. gag and pol encode polyproteins, each managing the capsid and replication. The pol region encodes enzymes necessary for viral replication, such as reverse transcriptase, protease and integrase. Depending on the virus, the genes may overlap or fuse into larger polyprotein chains. Some viruses contain additional genes. The lentivirus genus, the spumavirus genus, the HTLV / bovine leukemia virus (BLV) genus, and a newly introduced fish virus genus are retroviruses classified as complex. These viruses have genes called accessory genes, in addition to gag, pro, pol and env genes. Accessory genes are located between pol and env, downstream from the env, including the U3 region of LTR, or in the env and overlapping portions. While accessory genes have auxiliary roles, they also coordinate and regulate viral gene expression. In addition, some retroviruses may carry genes called oncogenes or onc genes from another class. Retroviruses with these genes (also called transforming viruses) are known for their ability to quickly cause tumors in animals and transform cells in culture into an oncogenic state.
The polyproteins are cleaved into smaller proteins each with their own function. The nucleotides encoding them are known as subgenes. |

|
|
|
|
| |
Multiplication
|
Multiplication
Multiplication (W)
When retroviruses have integrated their own genome into the germ line, their genome is passed on to a following generation. These endogenous retroviruses (ERVs), contrasted with exogenous ones, now make up 5–8% of the human genome. Most insertions have no known function and are often referred to as "junk DNA". However, many endogenous retroviruses play important roles in host biology, such as control of gene transcription, cell fusion during placental development in the course of the germination of an embryo, and resistance to exogenous retroviral infection. Endogenous retroviruses have also received special attention in the research of immunology-related pathologies, such as autoimmune diseases like multiple sclerosis, although endogenous retroviruses have not yet been proven to play any causal role in this class of disease.
While transcription was classically thought to occur only from DNA to RNA, reverse transcriptase transcribes RNA into DNA. The term "retro" in retrovirus refers to this reversal (making DNA from RNA) of the usual direction of transcription. It still obeys the central dogma of molecular biology, which states that information can be transferred from nucleic acid to nucleic acid but cannot be transferred back from protein to either protein or nucleic acid. Reverse transcriptase activity outside of retroviruses has been found in almost all eukaryotes, enabling the generation and insertion of new copies of retrotransposons into the host genome. These inserts are transcribed by enzymes of the host into new RNA molecules that enter the cytosol. Next, some of these RNA molecules are translated into viral proteins. The proteins encoded by the gag and pol genes are translated from genome-length mRNAs into Gag and Gag–Pol polyproteins. In example, for the gag gene; it is translated into molecules of the capsid protein, and for the pol gene; it is translated into molecules of reverse transcriptase. Retroviruses need a lot more amount of the Gag proteins than the Pol proteins and have developed advanced systems to synthesize the required amount of each. As an example, after Gag synthesis nearly 95 percent of the ribosomes terminate translation, while other ribosomes continue translation to synthesize Gag–Pol. In the rough endoplasmic reticulum glycosylation begins and the env gene is translated from spliced mRNAs in the rough endoplasmic reticulum, into molecules of the envelope protein. When the envelope protein molecules are carried to the Golgi complex, they are divided into surface glycoprotein and transmembrane glycoprotein by a host protease. These two glycoprotein products stay in close affiliation, and they are transported to the plasma membrane after further glycosylation.
It is important to note that a retrovirus must "bring" its own reverse transcriptase in its capsid, otherwise it is unable to utilize the enzymes of the infected cell to carry out the task, due to the unusual nature of producing DNA from RNA.
Industrial drugs that are designed as protease and reverse-transcriptase inhibitors are made such that they target specific sites and sequences within their respective enzymes. However these drugs can quickly become ineffective due to the fact that the gene sequences that code for the protease and the reverse transcriptase quickly mutate. These changes in bases cause specific codons and sites with the enzymes to change and thereby avoid drug targeting by losing the sites that the drug actually targets.
Because reverse transcription lacks the usual proofreading of DNA replication, a retrovirus mutates very often. This enables the virus to grow resistant to antiviral pharmaceuticals quickly, and impedes the development of effective vaccines and inhibitors for the retrovirus.
One difficulty faced with some retroviruses, such as the Moloney retrovirus, involves the requirement for cells to be actively dividing for transduction. As a result, cells such as neurons are very resistant to infection and transduction by retroviruses. This gives rise to a concern that insertional mutagenesis due to integration into the host genome might lead to cancer or leukemia. This is unlike Lentivirus, a genus of Retroviridae, which are able to integrate their RNA into the genome of non-dividing host cells. |
| |
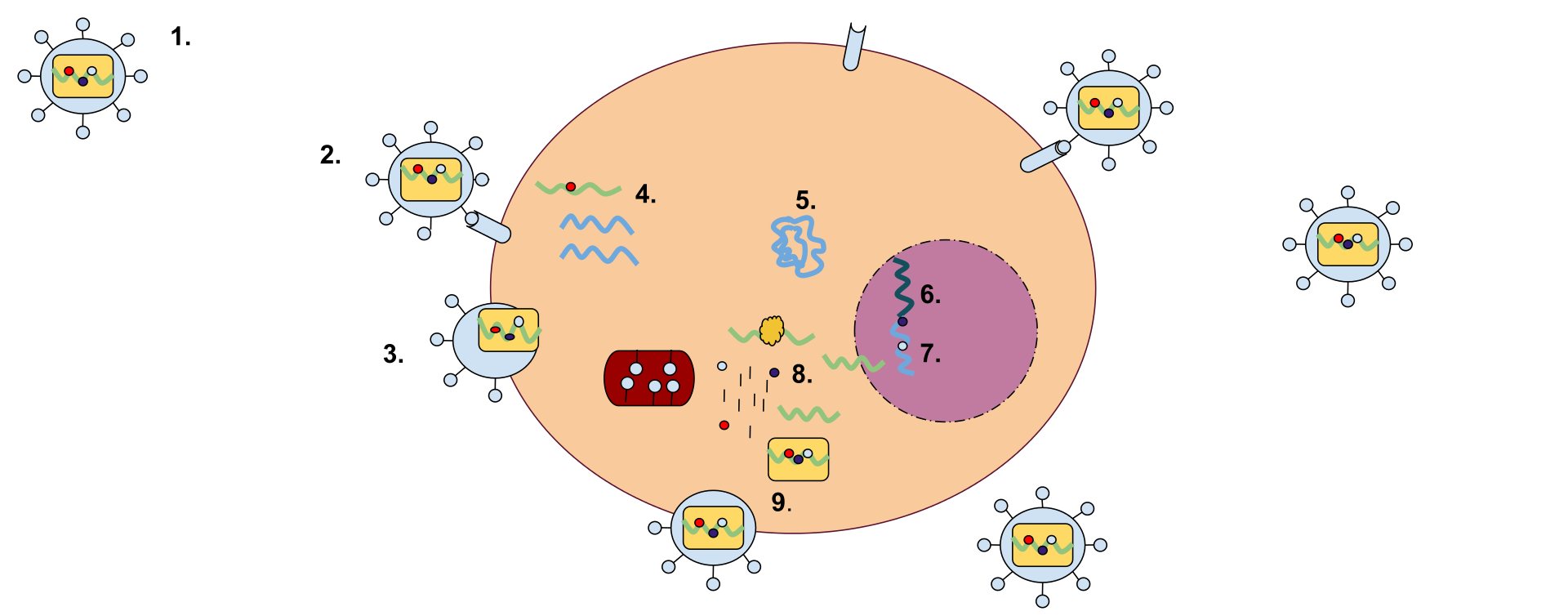
A retrovirus has a membrane containing glycoproteins, which are able to bind to a receptor protein on a host cell. There are two strands of RNA within the cell that have three enzymes: protease, reverse transcriptase, and integrase (1). The first step of replication is the binding of the glycoprotein to the receptor protein (2). Once these have been bound, the cell membrane degrades, becoming part of the host cell, and the RNA strands and enzymes enter the cell (3). Within the cell, reverse transcriptase creates a complementary strand of DNA from the retrovirus RNA and the RNA is degraded; this strand of DNA is known as cDNA (4). The cDNA is then replicated, and the two strands form a weak bond and enter the nucleus (5). Once in the nucleus, the DNA is integrated into the host cell's DNA with the help of integrase (6). This cell can either stay dormant, or RNA may be synthesized from the DNA and used to create the proteins for a new retrovirus (7). Ribosome units are used to translate the mRNA of the virus into the amino acid sequences which can be made into proteins in the rough endoplasmic reticulum. This step will also make viral enzymes and capsid proteins (8). Viral RNA will be made in the nucleus. These pieces are then gathered together and are pinched off of the cell membrane as a new retrovirus (9). |
|

|
|
|
|
Recombination
Recombination (W)
Two RNA genomes are packaged into each retrovirus particle, but, after an infection, each virus generates only one provirus. After infection, reverse transcription occurs and this process is accompanied by recombination. Recombination involves template strand switching between the two genome copies (copy choice recombination) during reverse transcription. From 5 to 14 recombination events per genome occur at each replication cycle. Genetic recombination appears to be necessary for maintaining genome integrity and as a repair mechanism for salvaging damaged genomes. |
|
|
|
| |
Transmission
|
|
| |
Provirus
|
Provirus
Provirus (W)
The DNA formed after reverse transcription (the provirus) is longer than the RNA genome because each of the terminals have the U3 - R - U5 sequences called long terminal repeat (LTR). Thus, 5' terminal has the extra U3 sequence, while the other terminal has the U5 sequence. LTRs are able to send signals for vital tasks to be carried out such as initiation of RNA production or management of the rate of transcription. This way, LTRs can control replication, hence, the entire progress of the viral cycle. Although located in the nucleus, the non-integrated retroviral cDNA is a very weak substrate for transcription. For this reason, an integrated provirus is a necessary for permanent and an effective expression of retroviral genes.
This DNA can be incorporated into host genome as a provirus that can be passed on to progeny cells. The retrovirus DNA is inserted at random into the host genome. Because of this, it can be inserted into oncogenes. In this way some retroviruses can convert normal cells into cancer cells. Some provirus remains latent in the cell for a long period of time before it is activated by the change in cell environment. |

|
|
|
|
| |
Early evolution
|
Early evolution
Early evolution (W)
Studies of retroviruses led to the first demonstrated synthesis of DNA from RNA templates, a fundamental mode for transferring genetic material that occurs in both eukaryotes and prokaryotes. It has been speculated that the RNA to DNA transcription processes used by retroviruses may have first caused DNA to be used as genetic material. In this model, the RNA world hypothesis, cellular organisms adopted the more chemically stable DNA when retroviruses evolved to create DNA from the RNA templates.
An estimate of the date of evolution of the foamy-like endogenous retroviruses placed the time of the most recent common ancestor at > 450 million years ago. |
|
|
|
| |
Gene therapy
|
Gene therapy
Gene therapy (W)
Gammaretroviral and lentiviral vectors for gene therapy have been developed that mediate stable genetic modification of treated cells by chromosomal integration of the transferred vector genomes. This technology is of use, not only for research purposes, but also for clinical gene therapy aiming at the long-term correction of genetic defects, e.g., in stem and progenitor cells. Retroviral vector particles with tropism for various target cells have been designed. Gammaretroviral and lentiviral vectors have so far been used in more than 300 clinical trials, addressing treatment options for various diseases. Retroviral mutations can be developed to make transgenic mouse models to study various cancers and their metastatic models. |
|
|
|
| |
Cancer
|
Cancer
Cancer (W)
Retroviruses that cause tumor growth include Rous sarcoma virus and Mouse mammary tumor virus. Cancer can be triggered by proto-oncogenes that were mistakenly incorporated into proviral DNA or by the disruption of cellular proto-oncogenes. Rous sarcoma virus contains the src gene that triggers tumor formation. Later it was found that a similar gene in cells is involved in cell signaling, which was most likely excised with the proviral DNA. Nontransforming viruses can randomly insert their DNA into proto-oncogenes, disrupting the expression of proteins that regulate the cell cycle. The promoter of the provirus DNA can also cause over expression of regulatory genes. Retroviruses can cause diseases such as cancer and immunodeficiency. If viral DNA is integrated into host chromosomes, it can lead to permanent infections. It is therefore important to discover the body's response to retroviruses. Especially exogenous retroviruses are associated with pathogenic diseases. For example, mice have mouse mammary tumor virus (MMTV), which is a retrovirus. This virus passes to newborn mice through mammary milk. The mice carrying the virus get mammary cancer because of the retrovirus when they are 6 months old. In addition, leukemia virus I (HTLV-1), found in human T cell, has been found in humans for many years. It is estimated that this retrovirus causes leukemia in the ages of 40 and 50.It has a replicable structure that can induce cancer. In addition to the usual gene sequence of retroviruses, HTLV-1 contains a fourth region, PX. This region encodes Tax, Rex, p12, p13 and p30 regulatory proteins. The Tax protein initiates the leukemic process and organizes the transcription of all viral genes in the integrated HTLV proviral DNA. |

|
|
|
|
| |
Classification
|
Classification
Classification (W)
|

Phylogeny of Retroviruses. |
|
|
|
|
Exogenous
Exogenous (W)
These are infectious RNA- or DNA-containing viruses which are transmitted from person to person.
Reverse-transcribing viruses fall into 2 groups of the Baltimore classification. |
|
|
|
Group VI viruses
Group VI viruses (W)
All members of Group VI use virally encoded reverse transcriptase, an RNA-dependent DNA polymerase, to produce DNA from the initial virion RNA genome. This DNA is often integrated into the host genome, as in the case of retroviruses and pseudoviruses, where it is replicated and transcribed by the host.
Group VI includes:
The family Retroviridae was previously divided into three subfamilies (Oncovirinae, Lentivirinae, and Spumavirinae), but are now divided into two: Orthoretrovirinae and Spumaretrovirinae. The term oncovirus is now commonly used to describe a cancer-causing virus. This family now includes the following genera:
Note that according to ICTV 2017, genus Spumavirus has been divided into five genera, and its former type species Simian foamy virus is now upgraded to genus Simiispumavirus with not less than 14 species, including new type species Eastern chimpanzee simian foamy virus.
|

|
|
|
|
Group VII viruses
Group VII viruses (W)
Both families in Group VII have DNA genomes contained within the invading virus particles. The DNA genome is transcribed into both mRNA, for use as a transcript in protein synthesis, and pre-genomic RNA, for use as the template during genome replication. Virally encoded reverse transcriptase uses the pre-genomic RNA as a template for the creation of genomic DNA.
Group VII includes:
The latter family is closely related to the newly proposed
whilst families Belpaoviridae, Metaviridae, Pseudoviridae, Retroviridae, and Caulimoviridae constitute the order Ortervirales. |
|
|
|
Endogenous
Endogenous (W)
Endogenous retroviruses are not formally included in this classification system, and are broadly classified into three classes, on the basis of relatedness to exogenous genera:
- Class I are most similar to the gammaretroviruses
- Class II are most similar to the betaretroviruses and alpharetroviruses
- Class III are most similar to the spumaviruses.
|
|
|
|
| |
Treatment
|
Treatment
Treatment (W)
Antiretroviral drugs are medications for the treatment of infection by retroviruses, primarily HIV. Different classes of antiretroviral drugs act on different stages of the HIV life cycle. Combination of several (typically three or four) antiretroviral drugs is known as highly active anti-retroviral therapy (HAART). |
|
|
|
| |
Treatment of veterinary retroviruses
|
Treatment of veterinary retroviruses
|
|
|

|
|
|
|
|
|

