Protein (B)
Protein (B)
Introduction
|
Introduction
Introduction (B)
Protein, highly complex substance that is present in all living organisms. Proteins are of great nutritional value and are directly involved in the chemical processes essential for life. The importance of proteins was recognized by chemists in the early 19th century, including Swedish chemist Jöns Jacob Berzelius, who in 1838 coined the term protein, a word derived from the Greek prōteios, meaning “holding first place.” Proteins are species-specific; that is, the proteins of one species differ from those of another species. They are also organ-specific; for instance, within a single organism, muscle proteins differ from those of the brain and liver. |
| |
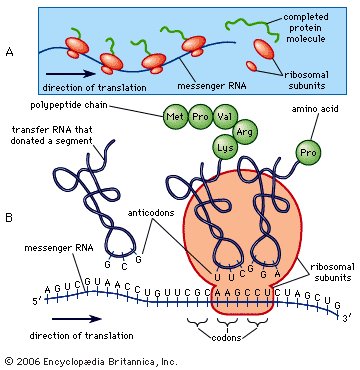
Synthesis of protein. |
|
|
|
| |
| A protein molecule is very large compared with molecules of sugar or salt and consists of many amino acids joined together to form long chains, much as beads are arranged on a string. There are about 20 different amino acids that occur naturally in proteins. Proteins of similar function have similar amino acid composition and sequence. Although it is not yet possible to explain all of the functions of a protein from its amino acid sequence, established correlations between structure and function can be attributed to the properties of the amino acids that compose proteins. |
| |
Peptide
The molecular structure of a peptide (a small protein) consists of a sequence of amino acids. |
|
|
|
| |
Plants can synthesize all of the amino acids; animals cannot, even though all of them are essential for life. Plants can grow in a medium containing inorganic nutrients that provide nitrogen, potassium, and other substances essential for growth. They utilize the carbon dioxide in the air during the process of photosynthesis to form organic compounds such as carbohydrates. Animals, however, must obtain organic nutrients from outside sources. Because the protein content of most plants is low, very large amounts of plant material are required by animals, such as ruminants (e.g., cows), that eat only plant material to meet their amino acid requirements. Nonruminant animals, including humans, obtain proteins principally from animals and their products—e.g., meat, milk, and eggs. The seeds of legumes are increasingly being used to prepare inexpensive protein-rich food (see human nutrition).
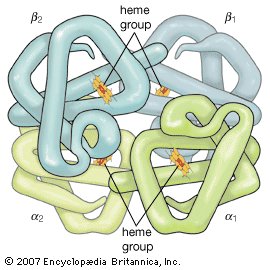
Hemoglobin
Hemoglobin is a protein made up of four polypeptide chains (α1, α2, β1, and β2). Each chain is attached to a heme group composed of porphyrin (an organic ringlike compound) attached to an iron atom. These iron-porphyrin complexes coordinate oxygen molecules reversibly, an ability directly related to the role of hemoglobin in oxygen transport in the blood. |
|
|
| |
|
The protein content of animal organs is usually much higher than that of the blood plasma. Muscles, for example, contain about 30 percent protein, the liver 20 to 30 percent, and red blood cells 30 percent. Higher percentages of protein are found in hair, bones, and other organs and tissues with a low water content. The quantity of free amino acids and peptides in animals is much smaller than the amount of protein; protein molecules are produced in cells by the stepwise alignment of amino acids and are released into the body fluids only after synthesis is complete.
The high protein content of some organs does not mean that the importance of proteins is related to their amount in an organism or tissue; on the contrary, some of the most important proteins, such as enzymes and hormones, occur in extremely small amounts. The importance of proteins is related principally to their function. All enzymes identified thus far are proteins. Enzymes, which are the catalysts of all metabolic reactions, enable an organism to build up the chemical substances necessary for life — proteins, nucleic acids, carbohydrates, and lipids — to convert them into other substances, and to degrade them. Life without enzymes is not possible. There are several protein hormones with important regulatory functions. In all vertebrates, the respiratory protein hemoglobin acts as oxygen carrier in the blood, transporting oxygen from the lung to body organs and tissues. A large group of structural proteins maintains and protects the structure of the animal body. |

|
|
|
|
| |
General Structure And Properties Of Proteins
|
The amino acid composition of proteins
The amino acid composition of proteins (B)
The common property of all proteins is that they consist of long chains of α-amino (alpha amino) acids. The general structure of α-amino acids is shown in . The α-amino acids are so called because the α-carbon atom in the molecule carries an amino group (―NH2); the α-carbon atom also carries a carboxyl group (―COOH). |
| |
|
In acidic solutions, when the pH is less than 4, the ―COO groups combine with hydrogen ions (H+) and are thus converted into the uncharged form (―COOH). In alkaline solutions, at pH above 9, the ammonium groups (―NH+3) lose a hydrogen ion and are converted into amino groups (―NH2). In the pH range between 4 and 8, amino acids carry both a positive and a negative charge and therefore do not migrate in an electrical field. Such structures have been designated as dipolar ions, or zwitterions (i.e., hybrid ions).
Although more than 100 amino acids occur in nature, particularly in plants, only 20 types are commonly found in most proteins. In protein molecules the α-amino acids are linked to each other by peptide bonds between the amino group of one amino acid and the carboxyl group of its neighbour. |
|
| The condensation (joining) of three amino acids yields the tripeptide. |
| |
|
| It is customary to write the structure of peptides in such a way that the free α-amino group (also called the N terminus of the peptide) is at the left side and the free carboxyl group (the C terminus) at the right side. Proteins are macromolecular polypeptides — i.e., very large molecules (macromolecules) composed of many peptide-bonded amino acids. Most of the common ones contain more than 100 amino acids linked to each other in a long peptide chain. The average molecular weight (based on the weight of a hydrogen atom as 1) of each amino acid is approximately 100 to 125; thus, the molecular weights of proteins are usually in the range of 10,000 to 100,000 daltons (one dalton is the weight of one hydrogen atom). The species-specificity and organ-specificity of proteins result from differences in the number and sequences of amino acids. Twenty different amino acids in a chain 100 amino acids long can be arranged in far more than 10100 ways (10100 is the number one followed by 100 zeroes). |

|
|
|
|
Structures of common amino acids
Structures of common amino acids (B)
The amino acids present in proteins differ from each other in the structure of their side (R) chains. The simplest amino acid is glycine, in which R is a hydrogen atom. In a number of amino acids, R represents straight or branched carbon chains. One of these amino acids is alanine, in which R is the methyl group (―CH3). Valine, leucine, and isoleucine, with longer R groups, complete the alkyl side-chain series. The alkyl side chains (R groups) of these amino acids are nonpolar; this means that they have no affinity for water but some affinity for each other. Although plants can form all of the alkyl amino acids, animals can synthesize only alanine and glycine; thus valine, leucine, and isoleucine must be supplied in the diet.
Two amino acids, each containing three carbon atoms, are derived from alanine; they are serine and cysteine. Serine contains an alcohol group (―CH2OH) instead of the methyl group of alanine, and cysteine contains a mercapto group (―CH2SH). Animals can synthesize serine but not cysteine or cystine. Cysteine occurs in proteins predominantly in its oxidized form (oxidation in this sense meaning the removal of hydrogen atoms), called cystine. Cystine consists of two cysteine molecules linked by the disulfide bond (―S―S―) that results when a hydrogen atom is removed from the mercapto group of each of the cysteines. Disulfide bonds are important in protein structure because they allow the linkage of two different parts of a protein molecule to—and thus the formation of loops in—the otherwise straight chains. Some proteins contain small amounts of cysteine with free sulfhydryl (―SH) groups. |
| |
|
Four amino acids, each consisting of four carbon atoms, occur in proteins; they are aspartic acid, asparagine, threonine, and methionine. Aspartic acid and asparagine, which occur in large amounts, can be synthesized by animals. Threonine and methionine cannot be synthesized and thus are essential amino acids; i.e., they must be supplied in the diet. Most proteins contain only small amounts of methionine.
Proteins also contain an amino acid with five carbon atoms (glutamic acid) and a secondary amine (in proline), which is a structure with the amino group (―NH2) bonded to the alkyl side chain, forming a ring. Glutamic acid and aspartic acid are dicarboxylic acids; that is, they have two carboxyl groups (―COOH). |
| |
|
| Glutamine is similar to asparagine in that both are the amides of their corresponding dicarboxylic acid forms; i.e., they have an amide group (―CONH2) in place of the carboxyl (―COOH) of the side chain. Glutamic acid and glutamine are abundant in most proteins; e.g., in plant proteins they sometimes comprise more than one-third of the amino acids present. Both glutamic acid and glutamine can be synthesized by animals. |
| |
Amino acid content of some proteins*
| amino acid |
protein |
| alpha-casein |
gliadin |
edestin |
collagen (ox hide) |
keratin (wool) |
myosin |
| *Number of gram molecules of amino acid per 100,000 grams of protein. |
| **The values for aspartic acid and glutamic acid include asparagine and glutamine, respectively. |
| ***Isoleucine plus leucine. |
| lysine |
60.9 |
4.45 |
19.9 |
27.4 |
6.2 |
85 |
| histidine |
18.7 |
11.7 |
18.6 |
4.5 |
19.7 |
15 |
| arginine |
24.7 |
15.7 |
99.2 |
47.1 |
56.9 |
41 |
| aspartic acid** |
63.1 |
10.1 |
99.4 |
51.9 |
51.5 |
85 |
| threonine |
41.2 |
17.6 |
31.2 |
19.3 |
55.9 |
41 |
| serine |
63.1 |
46.7 |
55.7 |
41.0 |
79.5 |
41 |
| glutamic acid** |
153.1 |
311.0 |
144.9 |
76.2 |
99.0 |
155 |
| proline |
71.3 |
117.8 |
32.9 |
125.2 |
58.3 |
22 |
| glycine |
37.3 |
— |
68.0 |
354.6 |
78.0 |
39 |
| alanine |
41.5 |
23.9 |
57.7 |
115.7 |
43.8 |
78 |
| half-cystine |
3.6 |
21.3 |
10.9 |
0.0 |
105.0 |
86 |
| valine |
53.8 |
22.7 |
54.6 |
21.4 |
46.6 |
42 |
| methionine |
16.8 |
11.3 |
16.4 |
6.5 |
4.0 |
22 |
| isoleucine |
48.8 |
90.8*** |
41.9 |
14.5 |
29.0 |
42 |
| leucine |
60.3 |
|
60.0 |
28.2 |
59.9 |
79 |
| tyrosine |
44.7 |
17.7 |
26.9 |
5.5 |
28.7 |
18 |
| phenylalanine |
27.9 |
39.0 |
38.4 |
13.9 |
22.4 |
27 |
| tryptophan |
7.8 |
3.2 |
6.6 |
0.0 |
9.6 |
— |
| hydroxyproline |
0.0 |
0.0 |
0.0 |
97.5 |
12.2 |
— |
| hydroxylysine |
— |
— |
— |
8.0 |
1.2 |
— |
| total |
839 |
765 |
883 |
1,058 |
863 |
832 |
| average residual weight |
119 |
131 |
113 |
95 |
117 |
120 |
|
| |
The amino acids proline and hydroxyproline occur in large amounts in collagen, the protein of the connective tissue of animals. Proline and hydroxyproline lack free amino (―NH2) groups because the amino group is enclosed in a ring structure with the side chain; they thus cannot exist in a zwitterion form. Although the nitrogen-containing group (>NH) of these amino acids can form a peptide bond with the carboxyl group of another amino acid, the bond so formed gives rise to a kink in the peptide chain; i.e., the ring structure alters the regular bond angle of normal peptide bonds.
Proteins usually are almost neutral molecules; that is, they have neither acidic nor basic properties. This means that the acidic carboxyl ( ―COO−) groups of aspartic and glutamic acid are about equal in number to the amino acids with basic side chains. Three such basic amino acids, each containing six carbon atoms, occur in proteins. The one with the simplest structure, lysine, is synthesized by plants but not by animals. Even some plants have a low lysine content. Arginine is found in all proteins; it occurs in particularly high amounts in the strongly basic protamines (simple proteins composed of relatively few amino acids) of fish sperm. The third basic amino acid is histidine. Both arginine and histidine can be synthesized by animals. Histidine is a weaker base than either lysine or arginine. The imidazole ring, a five-membered ring structure containing two nitrogen atoms in the side chain of histidine, acts as a buffer (i.e., a stabilizer of hydrogen ion concentration) by binding hydrogen ions (H+) to the nitrogen atoms of the imidazole ring. |
| |
|
| The remaining amino acids—phenylalanine, tyrosine, and tryptophan—have in common an aromatic structure; i.e., a benzene ring is present. These three amino acids are essential, and, while animals cannot synthesize the benzene ring itself, they can convert phenylalanine to tyrosine. |
| |
|
Because these amino acids contain benzene rings, they can absorb ultraviolet light at wavelengths between 270 and 290 nanometres (nm; 1 nanometre = 10−9 metre = 10 angstrom units). Phenylalanine absorbs very little ultraviolet light; tyrosine and tryptophan, however, absorb it strongly and are responsible for the absorption band most proteins exhibit at 280–290 nanometres. This absorption is often used to determine the quantity of protein present in protein samples.
Most proteins contain only the amino acids described above; however, other amino acids occur in proteins in small amounts. For example, the collagen found in connective tissue contains, in addition to hydroxyproline, small amounts of hydroxylysine. Other proteins contain some monomethyl-, dimethyl-, or trimethyllysine—i.e., lysine derivatives containing one, two, or three methyl groups (―CH3). The amount of these unusual amino acids in proteins, however, rarely exceeds 1 or 2 percent of the total amino acids. |

|
|
|
|
Physicochemical properties of the amino acids
Physicochemical properties of the amino acids (B)
The physicochemical properties of a protein are determined by the analogous properties of the amino acids in it.
The α-carbon atom of all amino acids, with the exception of glycine, is asymmetric; this means that four different chemical entities (atoms or groups of atoms) are attached to it. As a result, each of the amino acids, except glycine, can exist in two different spatial, or geometric, arrangements (i.e., isomers), which are mirror images akin to right and left hands.
These isomers exhibit the property of optical rotation. Optical rotation is the rotation of the plane of polarized light, which is composed of light waves that vibrate in one plane, or direction, only. Solutions of substances that rotate the plane of polarization are said to be optically active, and the degree of rotation is called the optical rotation of the solution. The direction in which the light is rotated is generally designed as plus, or d, for dextrorotatory (to the right), or as minus, or l, for levorotatory (to the left). Some amino acids are dextrorotatory, others are levorotatory. With the exception of a few small proteins (peptides) that occur in bacteria, the amino acids that occur in proteins are L-amino acids. |
| |
|
In bacteria, D-alanine and some other D-amino acids have been found as components of gramicidin and bacitracin. These peptides are toxic to other bacteria and are used in medicine as antibiotics. The D-alanine has also been found in some peptides of bacterial membranes.
In contrast to most organic acids and amines, the amino acids are insoluble in organic solvents. In aqueous solutions they are dipolar ions (zwitterions, or hybrid ions) that react with strong acids or bases in a way that leads to the neutralization of the negatively or positively charged ends, respectively. Because of their reactions with strong acids and strong bases, the amino acids act as buffers—stabilizers of hydrogen ion (H+) or hydroxide ion (OH−) concentrations. In fact, glycine is frequently used as a buffer in the pH range from 1 to 3 (acid solutions) and from 9 to 12 (basic solutions). In acid solutions, glycine has a positive charge and therefore migrates to the cathode (negative electrode of a direct-current electrical circuit with terminals in the solution). Its charge, however, is negative in alkaline solutions, in which it migrates to the anode (positive electrode). At pH 6.1 glycine does not migrate, because each molecule has one positive and one negative charge. The pH at which an amino acid does not migrate in an electrical field is called the isoelectric point. Most of the monoamino acids (i.e., those with only one amino group) have isoelectric points similar to that of glycine. The isoelectric points of aspartic and glutamic acids, however, are close to pH 3, and those of histidine, lysine, and arginine are at pH 7.6, 9.7, and 10.8, respectively. |

|
|
|
|
Amino acid sequence in protein molecules
Amino acid sequence in protein molecules (B)
Amino acid sequence in protein molecules
Since each protein molecule consists of a long chain of amino acid residues, linked to each other by peptide bonds, the hydrolytic cleavage of all peptide bonds is a prerequisite for the quantitative determination of the amino acid residues. Hydrolysis is most frequently accomplished by boiling the protein with concentrated hydrochloric acid. The quantitative determination of the amino acids is based on the discovery that amino acids can be separated from each other by chromatography on filter paper and made visible by spraying the paper with ninhydrin. The amino acids of the protein hydrolysate are separated from each other by passing the hydrolysate through a column of adsorbents, which adsorb the amino acids with different affinities and, on washing the column with buffer solutions, release them in a definite order. The amount of each of the amino acids can be determined by the intensity of the colour reaction with ninhydrin.
To obtain information about the sequence of the amino acid residues in the protein, the protein is degraded stepwise, one amino acid being split off in each step. This is accomplished by coupling the free α-amino group (―NH2) of the N-terminal amino acid with phenyl isothiocyanate; subsequent mild hydrolysis does not affect the peptide bonds. The procedure, called the Edman degradation, can be applied repeatedly; it thus reveals the sequence of the amino acids in the peptide chain.
Unavoidable small losses that occur during each step make it impossible to determine the sequence of more than about 30 to 50 amino acids by this procedure. For this reason the protein is usually first hydrolyzed by exposure to the enzyme trypsin, which cleaves only peptide bonds formed by the carboxyl groups of lysine and arginine. The Edman degradation is then applied to each of the few resulting peptides produced by the action of trypsin. Further information can be gained by hydrolyzing another portion of the protein with another enzyme, for instance with chymotrypsin, which splits predominantly peptide bonds formed by the amino acids tyrosine, phenylalanine, and tryptophan. The combination of results obtained with two or more different proteolytic (protein degrading) enzymes was first applied by English biochemist Frederick Sanger, and it enabled him to elucidate the amino acid sequence of insulin. The amino acid sequences of many other proteins subsequently were determined in the same manner. |

|
|
|
|
| |
Levels of structural organization in proteins
|
Primary structure
Primary structure (B)
Analytical and synthetic procedures reveal only the primary structure of the proteins—that is, the amino acid sequence of the peptide chains. They do not reveal information about the conformation (arrangement in space) of the peptide chain—that is, whether the peptide chain is present as a long straight thread or is irregularly coiled and folded into a globule. The configuration, or conformation, of a protein is determined by mutual attraction or repulsion of polar or nonpolar groups in the side chains (R groups) of the amino acids. The former have positive or negative charges in their side chains; the latter repel water but attract each other. Some parts of a peptide chain containing 100 to 200 amino acids may form a loop, or helix; others may be straight or form irregular coils.
The terms secondary, tertiary, and quaternary structure are frequently applied to the configuration of the peptide chain of a protein. A nomenclature committee of the International Union of Biochemistry (IUB) has defined these terms as follows: The primary structure of a protein is determined by its amino acid sequence without any regard for the arrangement of the peptide chain in space. The secondary structure is determined by the spatial arrangement of the main peptide chain without any regard for the conformation of side chains or other segments of the main chain. The tertiary structure is determined by both the side chains and other adjacent segments of the main chain, without regard for neighbouring peptide chains. Finally, the term quaternary structure is used for the arrangement of identical or different subunits of a large protein in which each subunit is a separate peptide chain. |

|
|
|
|
Secondary structure
Secondary structure (B)
The nitrogen and carbon atoms of a peptide chain cannot lie on a straight line, because of the magnitude of the bond angles between adjacent atoms of the chain; the bond angle is about 110°. Each of the nitrogen and carbon atoms can rotate to a certain extent, however, so that the chain has a limited flexibility. Because all of the amino acids, except glycine, are asymmetric L-amino acids, the peptide chain tends to assume an asymmetric helical shape; some of the fibrous proteins consist of elongated helices around a straight screw axis. Such structural features result from properties common to all peptide chains. The product of their effects is the secondary structure of the protein. |
|
|
|
Tertiary structure
Tertiary structure (B)
The tertiary structure is the product of the interaction between the side chains (R) of the amino acids composing the protein. Some of them contain positively or negatively charged groups, others are polar, and still others are nonpolar. The number of carbon atoms in the side chain varies from zero in glycine to nine in tryptophan. Positively and negatively charged side chains have the tendency to attract each other; side chains with identical charges repel each other. The bonds formed by the forces between the negatively charged side chains of aspartic or glutamic acid on the one hand, and the positively charged side chains of lysine or arginine on the other hand, are called salt bridges. Mutual attraction of adjacent peptide chains also results from the formation of numerous hydrogen bonds. |
| |
|
Hydrogen bonds form as a result of the attraction between the nitrogen-bound hydrogen atom (the imide hydrogen) and the unshared pair of electrons of the oxygen atom in the double bonded carbon–oxygen group (the carbonyl group). The result is a slight displacement of the imide hydrogen toward the oxygen atom of the carbonyl group. Although the hydrogen bond is much weaker than a covalent bond (i.e., the type of bond between two carbon atoms, which equally share the pair of bonding electrons between them), the large number of imide and carbonyl groups in peptide chains results in the formation of numerous hydrogen bonds. Another type of attraction is that between nonpolar side chains of valine, leucine, isoleucine, and phenylalanine; the attraction results in the displacement of water molecules and is called hydrophobic interaction.
In proteins rich in cystine, the conformation of the peptide chain is determined to a considerable extent by the disulfide bonds (―S―S―) of cystine. The halves of cystine may be located in different parts of the peptide chain and thus may form a loop closed by the disulfide bond. |
| |
|
| If the disulfide bond is reduced (i.e., hydrogen is added) to two sulfhydryl (―SH) groups, the tertiary structure of the protein undergoes a drastic change—closed loops are broken and adjacent disulfide-bonded peptide chains separate. |

|
|
|
|
Quaternary structure
Quaternary structure (B)
The nature of the quaternary structure is demonstrated by the structure of hemoglobin. Each molecule of human hemoglobin consists of four peptide chains, two α-chains and two β-chains; i.e., it is a tetramer. The four subunits are linked to each other by hydrogen bonds and hydrophobic interaction. Because the four subunits are so closely linked, the hemoglobin tetramer is called a molecule, even though no covalent bonds occur between the peptide chains of the four subunits. In other proteins, the subunits are bound to each other by covalent bonds (disulfide bridges).
The amino acid sequence of porcine proinsulin is shown below. The arrows indicate the direction from the N terminus of the β-chain (B) to the C terminus of the α-chain (A). |
| |
|
|
|
|
| |
The isolation and determination of proteins
|
The isolation and determination of proteins
The isolation and determination of proteins (B)
Animal material usually contains large amounts of protein and lipids and small amounts of carbohydrate; in plants, the bulk of the dry matter is usually carbohydrate. If it is necessary to determine the amount of protein in a mixture of animal foodstuffs, a sample is converted to ammonium salts by boiling with sulfuric acid and a suitable inorganic catalyst, such as copper sulfate ( Kjeldahl method). The method is based on the assumption that proteins contain 16 percent nitrogen, and that nonprotein nitrogen is present in very small amounts. The assumption is justified for most tissues from higher animals but not for insects and crustaceans, in which a considerable portion of the body nitrogen is present in the form of chitin, a carbohydrate. Large amounts of nonprotein nitrogen are also found in the sap of many plants. In such cases, the precise quantitative analyses are made after the proteins have been separated from other biological compounds.
Proteins are sensitive to heat, acids, bases, organic solvents, and radiation exposure; for this reason, the chemical methods employed to purify organic compounds cannot be applied to proteins. Salts and molecules of small size are removed from protein solutions by dialysis—i.e., by placing the solution into a sac of semipermeable material, such as cellulose or acetylcellulose, which will allow small molecules to pass through but not large protein molecules, and immersing the sac in water or a salt solution. Small molecules can also be removed either by passing the protein solution through a column of resin that adsorbs only the protein or by gel filtration. In gel filtration, the large protein molecules pass through the column, and the small molecules are adsorbed to the gel.
Groups of proteins are separated from each other by salting out—i.e., the stepwise addition of sodium sulfate or ammonium sulfate to a protein solution. Some proteins, called globulins, become insoluble and precipitate when the solution is half-saturated with ammonium sulfate or when its sodium sulfate content exceeds about 12 percent. Other proteins, the albumins, can be precipitated from the supernatant solution (i.e., the solution remaining after a precipitation has taken place) by saturation with ammonium sulfate. Water-soluble proteins can be obtained in a dry state by freeze-drying (lyophilization), in which the protein solution is deep-frozen by lowering the temperature below −15 °C (5 °F) and removing the water; the protein is obtained as a dry powder.
Most proteins are insoluble in boiling water and are denatured by it—i.e., irreversibly converted into an insoluble material. Heat denaturation cannot be used with connective tissue because the principal structural protein, collagen, is converted by boiling water into water-soluble gelatin.
Fractionation (separation into components) of a mixture of proteins of different molecular weight can be accomplished by gel filtration. The size of the proteins retained by the gel depends upon the properties of the gel. The proteins retained in the gel are removed from the column by solutions of a suitable concentration of salts and hydrogen ions.
Many proteins were originally obtained in crystalline form, but crystallinity is not proof of purity; many crystalline protein preparations contain other substances. Various tests are used to determine whether a protein preparation contains only one protein. The purity of a protein solution can be determined by such techniques as chromatography and gel filtration. In addition, a solution of pure protein will yield one peak when spun in a centrifuge at very high speeds (ultracentrifugation) and will migrate as a single band in electrophoresis (migration of the protein in an electrical field). After these methods and others (such as amino acid analysis) indicate that the protein solution is pure, it can be considered so. Because chromatography, ultracentrifugation, and electrophoresis cannot be applied to insoluble proteins, little is known about them; they may be mixtures of many similar proteins.
Very small (microheterogeneous) differences in some of the apparently pure proteins are known to occur. They are differences in the amino acid composition of otherwise identical proteins and are transmitted from generation to generation; i.e., they are genetically determined. For example, some humans have two hemoglobins, hemoglobin A and hemoglobin S, which differ in one amino acid at a specific site in the molecule. In hemoglobin A the site is occupied by glutamic acid and in hemoglobin S by valine. Refinement of the techniques of protein analysis has resulted in the discovery of other instances of microheterogeneity.
The quantity of a pure protein can be determined by weighing or by measuring the ultraviolet absorbancy at 280 nanometres. The absorbency at 280 nanometres depends on the content of tyrosine and tryptophan in the protein. Sometimes the slightly less sensitive biuret reaction, a purple colour given by alkaline protein solutions upon the addition of copper sulfate, is used; its intensity depends only on the number of peptide bonds per gram, which is similar in all proteins. |

|
|
|
|
| |
Physicochemical properties of proteins
|
The molecular weight of proteins
The molecular weight of proteins (B)
The molecular weight of proteins cannot be determined by the methods of classical chemistry (e.g., freezing-point depression), because they require solutions of a higher concentration of protein than can be prepared.
If a protein contains only one molecule of one of the amino acids or one atom of iron, copper, or another element, the minimum molecular weight of the protein or a subunit can be calculated; for example, the protein myoglobin contains 0.34 gram of iron in 100 grams of protein. The atomic weight of iron is 56; thus the minimum molecular weight of myoglobin is (56 × 100)/0.34 = about 16,500. Direct measurements of the molecular weight of myoglobin yield the same value. The molecular weight of hemoglobin, however, which also contains 0.34 percent iron, has been found to be 66,000 or 4 × 16,500; thus hemoglobin contains four atoms of iron.
The method most frequently used to determine the molecular weight of proteins is ultracentrifugation—i.e., spinning in a centrifuge at velocities up to about 60,000 revolutions per minute. Centrifugal forces of more than 200,000 times the gravitational force on the surface of Earth are achieved at such velocities. The first ultracentrifuges, built in 1920, were used to determine the molecular weight of proteins. The molecular weights of a large number of proteins have been determined. Most consist of several subunits, the molecular weight of which is usually less than 100,000 and frequently ranges from 20,000 to 30,000. Proteins of very high molecular weights are found among hemocyanins, the copper-containing respiratory proteins of invertebrates; some range as high as several million. Although there is no definite lower limit for the molecular weight of proteins, short amino acid sequences are usually called peptides. |

|
|
|
|
The shape of protein molecules
The shape of protein molecules (B)
|
|
| |
|
| X-ray diffraction pattern of a crystallized enzyme. |
|
| |
|
In the technique of X-ray diffraction, the X-rays are allowed to strike a protein crystal. The X-rays, diffracted (bent) by the crystal, impinge on a photographic plate, forming a pattern of spots. This method reveals that peptide chains can assume very complicated, apparently irregular shapes. Two extremes in shape include the closely folded structure of the globular proteins and the elongated, unidimensional structure of the threadlike fibrous proteins; both were recognized many years before the technique of X-ray diffraction was developed. Solutions of fibrous proteins are extremely viscous (i.e., sticky); those of the globular proteins have low viscosity (i.e., they flow easily). A 5 percent solution of a globular protein—ovalbumin, for example—easily flows through a narrow glass tube; a 5 percent solution of gelatin, a fibrous protein, however, does not flow through the tube, because it is liquid only at high temperatures and solidifies at room temperature. Even solutions containing only 1 or 2 percent of gelatin are highly viscous and flow through a narrow tube either very slowly or only under pressure. |
|
|
| |
|
| Figure 2: Flow birefringence. Orientation of elongated, rodlike macromolecules (A) in resting solution, or (B) during flow through a horizontal tube. |
|
| |
|
The elongated peptide chains of the fibrous proteins can be imagined to become entangled not only mechanically but also by mutual attraction of their side chains, and in this way they incorporate large amounts of water. Most of the hydrophilic (water-attracting) groups of the globular proteins, however, lie on the surface of the molecules, and, as a result, globular proteins incorporate only a few water molecules. If a solution of a fibrous protein flows through a narrow tube, the elongated molecules become oriented parallel to the direction of the flow, and the solution thus becomes birefringent like a crystal; i.e., it splits a light ray into two components that travel at different velocities and are polarized at right angles to each other. Globular proteins do not show this phenomenon, which is called flow birefringence. Solutions of myosin, the contractile protein of muscles, show very high flow birefringence; other proteins with very high flow birefringence include solutions of fibrinogen, the clotting material of blood plasma, and solutions of tobacco mosaic virus. The gamma-globulins of the blood plasma show low flow birefringence, and none can be observed in solutions of serum albumin and ovalbumin. |

|
|
|
|
Hydration of proteins
Hydration of proteins (B)
When dry proteins are exposed to air of high water content, they rapidly bind water up to a maximum quantity, which differs for different proteins; usually it is 10 to 20 percent of the weight of the protein. The hydrophilic groups of a protein are chiefly the positively charged groups in the side chains of lysine and arginine and the negatively charged groups of aspartic and glutamic acid. Hydration (i.e., the binding of water) may also occur at the hydroxyl (―OH) groups of serine and threonine or at the amide (―CONH2) groups of asparagine and glutamine.
The binding of water molecules to either charged or polar (partly charged) groups is explained by the dipolar structure of the water molecule; that is, the two positively charged hydrogen atoms form an angle of about 105°, with the negatively charged oxygen atom at the apex. The centre of the positive charges is located between the two hydrogen atoms; the centre of the negative charge of the oxygen atom is at the apex of the angle. The negative pole of the dipolar water molecule binds to positively charged groups; the positive pole binds negatively charged ones. The negative pole of the water molecule also binds to the hydroxyl and amino groups of the protein.
The water of hydration is essential to the structure of protein crystals; when they are completely dehydrated, the crystalline structure disintegrates. In some proteins this process is accompanied by denaturation and loss of the biological function.
In aqueous solutions, proteins bind some of the water molecules very firmly; others are either very loosely bound or form islands of water molecules between loops of folded peptide chains. Because the water molecules in such an island are thought to be oriented as in ice, which is crystalline water, the islands of water in proteins are called icebergs. Water molecules may also form bridges between the carbonyl and imino groups of adjacent peptide chains, resulting in structures similar to those of the pleated sheet but with a water molecule in the position of the hydrogen bonds of that configuration. The extent of hydration of protein molecules in aqueous solutions is important, because some of the methods used to determine the molecular weight of proteins yield the molecular weight of the hydrated protein. The amount of water bound to one gram of a globular protein in solution varies from 0.2 to 0.5 gram. Much larger amounts of water are mechanically immobilized between the elongated peptide chains of fibrous proteins; for example, one gram of gelatin can immobilize at room temperature 25 to 30 grams of water.
Hydration of proteins is necessary for their solubility in water. If the water of hydration of a protein dissolved in water is reduced by the addition of a salt such as ammonium sulfate, the protein is no longer soluble and is salted out, or precipitated. The salting-out process is reversible because the protein is not denatured (i.e., irreversibly converted to an insoluble material) by the addition of such salts as sodium chloride, sodium sulfate, or ammonium sulfate. Some globulins, called euglobulins, are insoluble in water in the absence of salts; their insolubility is attributed to the mutual interaction of polar groups on the surface of adjacent molecules, a process that results in the formation of large aggregates of molecules. Addition of small amounts of salt causes the euglobulins to become soluble. This process, called salting in, results from a combination between anions (negatively charged ions) and cations (positively charged ions) of the salt and positively and negatively charged side chains of the euglobulins. The combination prevents the aggregation of euglobulin molecules by preventing the formation of salt bridges between them. The addition of more sodium or ammonium sulfate causes the euglobulins to salt out again and to precipitate. |

|
|
|
|
Electrochemistry of proteins
Electrochemistry of proteins (B)
Because the α-amino group and α-carboxyl group of amino acids are converted into peptide bonds in the protein molecule, there is only one α-amino group (at the N terminus) and one α-carboxyl group (at the C terminus) in a given protein molecule. The electrochemical character of a protein is affected very little by these two groups. Of importance, however, are the numerous positively charged ammonium groups (―NH3+) of lysine and arginine and the negatively charged carboxyl groups (―COO−) of aspartic acid and glutamic acid. In most proteins, the number of positively and negatively charged groups varies from 10 to 20 per 100 amino acids.
When measured volumes of hydrochloric acid are added to a solution of protein in salt-free water, the pH decreases in proportion to the amount of hydrogen ions added until it is about 4. Further addition of acid causes much less decrease in pH because the protein acts as a buffer at pH values of 3 to 4. The reaction that takes place in this pH range is the protonation of the carboxyl group—i.e., the conversion of ―COO− into ―COOH. Electrometric titration of an isoelectric protein with potassium hydroxide causes a very slow increase in pH and a weak buffering action of the protein at pH 7; a very strong buffering action occurs in the pH range from 9 to 10. The buffering action at pH 7, which is caused by loss of protons (positively charged hydrogen) from the imidazolium groups (i.e., the five-member ring structure in the side chain) of histidine, is weak because the histidine content of proteins is usually low. The much stronger buffering action at pH values from 9 to 10 is caused by the loss of protons from the hydroxyl group of tyrosine and from the ammonium groups of lysine. Finally, protons are lost from the guanidinium groups (i.e., the nitrogen-containing terminal portion of the arginine side chains) of arginine at pH 12. Electrometric titrations of proteins yield similar curves. Electrometric titration makes possible the determination of the approximate number of carboxyl groups, ammonium groups, histidines, and tyrosines per molecule of protein. |
| |
|
|
| |
|
| Figure 3: Electrometric titration of glycine. |
|
|
| |
| Electrophoresis
The positively and negatively charged side chains of proteins cause them to behave like amino acids in an electrical field; that is, they migrate during electrophoresis at low pH values to the cathode (negative terminal) and at high pH values to the anode (positive terminal). The isoelectric point, the pH value at which the protein molecule does not migrate, is in the range of pH 5 to 7 for many proteins. Proteins such as lysozyme, cytochrome c, histone, and others rich in lysine and arginine, however, have isoelectric points in the pH range between 8 and 10. The isoelectric point of pepsin, which contains very few basic amino acids, is close to 1. |
| |
Number of amino acids per protein molecule
| amino acid |
protein* |
| Cyto |
Hb alpha |
Hb beta |
RNase |
Lys |
Chgen |
Fdox |
| *Cyto = human cytochrome c; Hb alpha = human hemoglobin A, alpha-chain; Hb beta = human hemoglobin A, beta-chain; RNase = bovine ribonuclease; Lys = chicken lysozyme; Chgen = bovine chymotrypsinogen; Fdox = spinach ferredoxin. |
| **The values recorded for aspartic acid and glutamic acid include asparagine and glutamine, respectively. |
| lysine |
18 |
11 |
11 |
10 |
6 |
14 |
4 |
| histidine |
3 |
10 |
9 |
4 |
1 |
2 |
1 |
| arginine |
2 |
3 |
3 |
4 |
11 |
4 |
1 |
| aspartic acid** |
8 |
12 |
13 |
15 |
21 |
23 |
13 |
| threonine |
7 |
9 |
7 |
10 |
7 |
23 |
8 |
| serine |
2 |
11 |
5 |
15 |
10 |
28 |
7 |
| glutamic acid** |
10 |
5 |
11 |
12 |
5 |
15 |
13 |
| proline |
4 |
7 |
7 |
4 |
2 |
9 |
4 |
| glycine |
13 |
7 |
13 |
3 |
12 |
23 |
6 |
| alanine |
6 |
21 |
15 |
12 |
12 |
22 |
9 |
| half-cystine |
2 |
1 |
2 |
8 |
8 |
10 |
5 |
| valine |
3 |
13 |
18 |
9 |
6 |
23 |
7 |
| methionine |
3 |
2 |
1 |
4 |
2 |
2 |
0 |
| isoleucine |
8 |
0 |
0 |
3 |
6 |
10 |
4 |
| leucine |
6 |
18 |
18 |
2 |
8 |
19 |
8 |
| tyrosine |
5 |
3 |
3 |
6 |
3 |
4 |
4 |
| phenylalanine |
3 |
7 |
8 |
3 |
3 |
6 |
2 |
| tryptophan |
1 |
1 |
2 |
0 |
6 |
8 |
1 |
| total |
104 |
141 |
146 |
124 |
129 |
245 |
97 |
|
| |
|
|
| |
|
Two-dimensional gel electrophoresis
In two-dimensional gel electrophoresis, proteins are separated based on charge and size. Approaches commonly employed include isoelectric focusing (IEF) sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) and immobilized pH gradient (IPG-Dalt) SDS-PAGE. |
|
|
| |
| Free-boundary electrophoresis, the original method of determining electrophoretic migration, has been replaced in many instances by zone electrophoresis, in which the protein is placed in either a gel of starch, agar, or polyacrylamide or in a porous medium such as paper or cellulose acetate. The migration of hemoglobin and other coloured proteins can be followed visually. Colourless proteins are made visible after the completion of electrophoresis by staining them with a suitable dye. |

|
|
|
|
| |
Conformation of globular proteins
|
Results of X-ray diffraction studies
Results of X-ray diffraction studies (B)
Most knowledge concerning secondary and tertiary structure of globular proteins has been obtained by the examination of their crystals using X-ray diffraction. In this technique, X-rays are allowed to strike the crystal; the X-rays are diffracted by the crystal and impinge on a photographic plate, forming a pattern of spots. The measured intensity of the diffraction pattern, as recorded on a photographic film, depends particularly on the electron density of the atoms in the protein crystal. This density is lowest in hydrogen atoms, and they do not give a visible diffraction pattern. Although carbon, oxygen, and nitrogen atoms yield visible diffraction patterns, they are present in such great number—about 700 or 800 per 100 amino acids—that the resolution of the structure of a protein containing more than 100 amino acids is almost impossible. Resolution is considerably improved by substituting into the side chains of certain amino acids very heavy atoms, particularly those of heavy metals. Mercury ions, for example, bind to the sulfhydryl (―SH) groups of cysteine. Platinum chloride has been used in other proteins. In the iron-containing proteins, the iron atom already in the molecule is adequate.
Although the X-ray diffraction technique cannot resolve the complete three-dimensional conformation (that is, the secondary and tertiary structure of the peptide chain), complete resolution has been obtained by combination of the results of X-ray diffraction with those of amino acid sequence analysis. In this way the complete conformation of such proteins as myoglobin, chymotrypsinogen, lysozyme, and ribonuclease has been resolved.
The X-ray diffraction method has revealed regular structural arrangements in proteins; one is an extended form of antiparallel peptide chains that are linked to each other by hydrogen bonds between the carbonyl and imino groups. This conformation, called the pleated sheet, or β-structure, is found in some fibrous proteins. Short strands of the β-structure have also been detected in some globular proteins.
|
|
| |
|
Protein structure; α-helix
The α-helix in the structural arrangement of a protein. |
|
| |
|
A second important structural arrangement is the α-helix; it is formed by a sequence of amino acids wound around a straight axis in either a right-handed or a left-handed spiral. Each turn of the helix corresponds to a distance of 5.4 angstroms (= 0.54 nanometre) in the direction of the screw axis and contains 3.7 amino acids. Hence, the length of the α-helix per amino acid residue is 5.4 divided by 3.7, or 1.5 angstroms (1 angstrom = 0.1 nanometre). The stability of the α-helix is maintained by hydrogen bonds between the carbonyl and imino groups of neighbouring turns of the helix. It was once thought, based on data from analyses of the myoglobin molecule, more than half of which consists of α-helices, that the α-helix is the predominant structural element of the globular proteins; it is now known that myoglobin is exceptional in this respect. The other globular proteins for which the structures have been resolved by X-ray diffraction contain only small regions of α-helix. In most of them the peptide chains are folded in an apparently random fashion, for which the term random coil has been used. The term is misleading, however, because the folding is not random; rather, it is dictated by the primary structure and modified by the secondary and tertiary structures.
The first proteins for which the internal structures were completely resolved are the iron-containing proteins myoglobin and hemoglobin. The investigation of the hydrated crystals of these proteins by Austrian-born British biochemist Max Perutz and British biochemist John C. Kendrew, who won the 1962 Nobel Prize for Chemistry for their work, revealed that the folding of the peptide chains is so tight that most of the water is displaced from the centre of the globular molecules. The amino acids that carry the ammonium (―NH3+) and carboxyl (―COO−) groups were found to be shifted to the surface of the globular molecules, and the nonpolar amino acids were found to be concentrated in the interior. |
| |
|
|
| |
|
Lysozyme; protein conformation
The simplified structure of lysozyme from hen's egg white has a single peptide chain of 129 amino acids. The amino acid residues are numbered from the terminal α group (N) to the terminal carboxyl group (C). Circles indicate every fifth residue, and every tenth residue is numbered. Broken lines indicate the four disulfide bridges. Alpha-helices are visible in the ranges 25 to 35, 90 to 100, and 120 to 125. |
|
| |
|
|

|
|
|
|
Other approaches to the determination of protein structure
Other approaches to the determination of protein structure (B)
None of the several other physical methods that have been used to obtain information on the secondary and tertiary structure of proteins provides as much direct information as the X-ray diffraction technique. Most of the techniques, however, are much simpler than X-ray diffraction, which requires, for the resolution of the structure of one protein, many years of work and equipment such as electronic computers. Some of the simpler techniques are based on the optical properties of proteins—refractivity, absorption of light of different wavelengths, rotation of the plane polarized light at different wavelengths, and luminescence.
Spectrophotometric behaviour
Spectrophotometry of protein solutions (the measurement of the degree of absorbance of light by a protein within a specified wavelength) is useful within the range of visible light only with proteins that contain coloured prosthetic groups (the nonprotein components). Examples of such proteins include the red heme proteins of the blood, the purple pigments of the retina of the eye, green and yellow proteins that contain bile pigments, blue copper-containing proteins, and dark brown proteins called melanins. Peptide bonds, because of their carbonyl groups, absorb light energy at very short wavelengths (185–200 nanometres). The aromatic rings of phenylalanine, tyrosine, and tryptophan, however, absorb ultraviolet light between wavelengths of 280 and 290 nanometres. The absorbance of ultraviolet light by tryptophan is greatest, that of tyrosine is less, and that of phenylalanine is least. If the tyrosine or tryptophan content of the protein is known, therefore, the concentration of the protein solution can be determined by measuring its absorbance between 280 and 290 nanometres.
It will be recalled that the amino acids, with the exception of glycine, exhibit optical activity (rotation of the plane of polarized light; see above Physicochemical properties of the amino acids). It is not surprising, therefore, that proteins also are optically active. They are usually levorotatory (i.e., they rotate the plane of polarization to the left) when polarized light of wavelengths in the visible range is used. Although the specific rotation (a function of the concentration of a protein solution and the distance the light travels in it) of most L-amino acids varies from −30° tο +30°, the amino acid cystine has a specific rotation of approximately −300°. Although the optical rotation of a protein depends on all of the amino acids of which it is composed, the most important ones are cystine and the aromatic amino acids phenylalanine, tyrosine, and tryptophan. The contribution of the other amino acids to the optical activity of a protein is negligibly small.
Chemical reactivity of proteins
Information on the internal structure of proteins can be obtained with chemical methods that reveal whether certain groups are present on the surface of the protein molecule and thus able to react or whether they are buried inside the closely folded peptide chains and thus are unable to react. The chemical reagents used in such investigations must be mild ones that do not affect the structure of the protein.
The reactivity of tyrosine is of special interest. It has been found, for example, that only three of the six tyrosines found in the naturally occurring enzyme ribonuclease can be iodinated (i.e., reacted to accept an iodine atom). Enzyme-catalyzed breakdown of iodinated ribonuclease is used to identify the peptides in which the iodinated tyrosines are present. The three tyrosines that can be iodinated lie on the surface of ribonuclease; the others, assumed to be inaccessible, are said to be buried in the molecule. Tyrosine can also be identified by using other techniques—e.g., treatment with diazonium compounds or tetranitromethane. Because the compounds formed are coloured, they can easily be detected when the protein is broken down with enzymes.
Cysteine can be detected by coupling with compounds such as iodoacetic acid or iodoacetamide; the reaction results in the formation of carboxymethylcysteine or carbamidomethylcysteine, which can be detected by amino acid determination of the peptides containing them. The imidazole groups of certain histidines can also be located by coupling with the same reagents under different conditions. Unfortunately, few other amino acids can be labelled without changes in the secondary and tertiary structure of the protein.
|

|
|
|
|
Association of protein subunits
Association of protein subunits (B)
Many proteins with molecular weights of more than 50,000 occur in aqueous solutions as complexes: dimers, tetramers, and higher polymers—i.e., as chains of two, four, or more repeating basic structural units. The subunits, which are called monomers or protomers, usually are present as an even number. Less than 10 percent of the polymers have been found to have an odd number of monomers. The arrangement of the subunits is thought to be regular and may be cyclic, cubic, or tetrahedral. Some of the small proteins also contain subunits. Insulin, for example, with a molecular weight of about 6,000, consists of two peptide chains linked to each other by disulfide bridges (―S―S―). Similar interchain disulfide bonds have been found in the immunoglobulins. In other proteins, hydrogen bonds and hydrophobic bonds (resulting from the interaction between the amino acid side chains of valine, leucine, isoleucine, and phenylalanine) cause the formation of aggregates of the subunits. The subunits of some proteins are identical; those of others differ. Hemoglobin is a tetramer consisting of two α-chains and two β-chains. |

|
|
|
|
Protein denaturation
Protein denaturation (B)
When a solution of a protein is boiled, the protein frequently becomes insoluble—i.e., it is denatured—and remains insoluble even when the solution is cooled. The denaturation of the proteins of egg white by heat—as when boiling an egg—is an example of irreversible denaturation. The denatured protein has the same primary structure as the original, or native, protein. The weak forces between charged groups and the weaker forces of mutual attraction of nonpolar groups are disrupted at elevated temperatures, however; as a result, the tertiary structure of the protein is lost. In some instances the original structure of the protein can be regenerated; the process is called renaturation.
Denaturation can be brought about in various ways. Proteins are denatured by treatment with alkaline or acid, oxidizing or reducing agents, and certain organic solvents. Interesting among denaturing agents are those that affect the secondary and tertiary structure without affecting the primary structure. The agents most frequently used for this purpose are urea and guanidinium chloride. These molecules, because of their high affinity for peptide bonds, break the hydrogen bonds and the salt bridges between positive and negative side chains, thereby abolishing the tertiary structure of the peptide chain. When denaturing agents are removed from a protein solution, the native protein re-forms in many cases. Denaturation can also be accomplished by reduction of the disulfide bonds of cystine—i.e., conversion of the disulfide bond (―S―S―) to two sulfhydryl groups (―SH). This, of course, results in the formation of two cysteines. Reoxidation of the cysteines by exposure to air sometimes regenerates the native protein. In other cases, however, the wrong cysteines become bound to each other, resulting in a different protein. Finally, denaturation can also be accomplished by exposing proteins to organic solvents such as ethanol or acetone. It is believed that the organic solvents interfere with the mutual attraction of nonpolar groups.
Some of the smaller proteins, however, are extremely stable, even against heat; for example, solutions of ribonuclease can be exposed for short periods of time to temperatures of 90 °C (194 °F) without undergoing significant denaturation. Denaturation does not involve identical changes in protein molecules. A common property of denatured proteins, however, is the loss of biological activity—e.g., the ability to act as enzymes or hormones.
Although denaturation had long been considered an all-or-none reaction, it is now thought that many intermediary states exist between native and denatured protein. In some instances, however, the breaking of a key bond could be followed by the complete breakdown of the conformation of the native protein.
Although many native proteins are resistant to the action of the enzyme trypsin, which breaks down proteins during digestion, they are hydrolyzed by the same enzyme after denaturation. The peptide bonds that can be split by trypsin are inaccessible in the native proteins but become accessible during denaturation. Similarly, denatured proteins give more intense colour reactions for tyrosine, histidine, and arginine than do the same proteins in the native state. The increased accessibility of reactive groups of denatured proteins is attributed to an unfolding of the peptide chains.
If denaturation can be brought about easily and if renaturation is difficult, how is the native conformation of globular proteins maintained in living organisms, in which they are produced stepwise, by incorporation of one amino acid at a time? Experiments on the biosynthesis of proteins from amino acids containing radioactive carbon or heavy hydrogen reveal that the protein molecule grows stepwise from the N terminus to the C terminus; in each step a single amino acid residue is incorporated. As soon as the growing peptide chain contains six or seven amino acid residues, the side chains interact with each other and thus cause deviations from the straight or β-chain configuration. Depending on the nature of the side chains, this may result in the formation of an α-helix or of loops closed by hydrogen bonds or disulfide bridges. The final conformation is probably frozen when the peptide chain attains a length of 50 or more amino acid residues. |

|
|
|
|
Conformation of proteins in interfaces
Conformation of proteins in interfaces (B)
Like many other substances with both hydrophilic and hydrophobic groups, soluble proteins tend to migrate into the interface between air and water or oil and water; the term oil here means a hydrophobic liquid such as benzene or xylene. Within the interface, proteins spread, forming thin films. Measurements of the surface tension, or interfacial tension, of such films indicate that tension is reduced by the protein film. Proteins, when forming an interfacial film, are present as a monomolecular layer—i.e., a layer one molecule in height. Although it was once thought that globular protein molecules unfold completely in the interface, it has now been established that many proteins can be recovered from films in the native state. The application of lateral pressure on a protein film causes it to increase in thickness and finally to form a layer with a height corresponding to the diameter of the native protein molecule. Protein molecules in an interface, because of Brownian motions (molecular vibrations), occupy much more space than do those in the film after the application of pressure. The Brownian motion of compressed molecules is limited to the two dimensions of the interface, since the protein molecules cannot move upward or downward.
The motion of protein molecules at the air–water interface has been used to determine the molecular weight of proteins. The technique involves measuring the force exerted by the protein layer on a barrier.
When a protein solution is vigorously shaken in air, it forms a foam, because the soluble proteins migrate into the air–water interface and persist there, preventing or slowing the reconversion of the foam into a homogeneous solution. Some of the unstable, easily modified proteins are denatured when spread in the air–water interface. The formation of a permanent foam when egg white is vigorously stirred is an example of irreversible denaturation by spreading in a surface. |

|
|
|
|
| |
Classification Of Proteins
|
Classification by solubility
Classification by solubility (B)
Collagen molecule. |
|
|
| |
|
After two German chemists, Emil Fischer and Franz Hofmeister, independently stated in 1902 that proteins are essentially polypeptides consisting of many amino acids, an attempt was made to classify proteins according to their chemical and physical properties, because the biological function of proteins had not yet been established. (The protein character of enzymes was not proved until the 1920s.) Proteins were classified primarily according to their solubility in a number of solvents. This classification is no longer satisfactory, however, because proteins of quite different structure and function sometimes have similar solubilities; conversely, proteins of the same function and similar structure sometimes have different solubilities. The terms associated with the old classification, however, are still widely used. They are defined below.
Albumins are proteins that are soluble in water and in water half-saturated with ammonium sulfate. On the other hand, globulins are salted out (i.e., precipitated) by half-saturation with ammonium sulfate. Globulins that are soluble in salt-free water are called pseudoglobulins; those insoluble in salt-free water are euglobulins. Both prolamins and glutelins, which are plant proteins, are insoluble in water; the prolamins dissolve in 50 to 80 percent ethanol, the glutelins in acidified or alkaline solution. The term protamine is used for a number of proteins in fish sperm that consist of approximately 80 percent arginine and therefore are strongly alkaline. Histones, which are less alkaline, apparently occur only in cell nuclei, where they are bound to nucleic acids. The term scleroproteins has been used for the insoluble proteins of animal organs. They include keratin, the insoluble protein of certain epithelial tissues such as the skin or hair, and collagen, the protein of the connective tissue. A large group of proteins has been called conjugated proteins, because they are complex molecules of protein consisting of protein and nonprotein moieties. The nonprotein portion is called the prosthetic group. Conjugated proteins can be subdivided into mucoproteins, which, in addition to protein, contain carbohydrate; lipoproteins, which contain lipids; phosphoproteins, which are rich in phosphate; chromoproteins, which contain pigments such as iron-porphyrins, carotenoids, bile pigments, and melanin; and finally, nucleoproteins, which contain nucleic acid. |
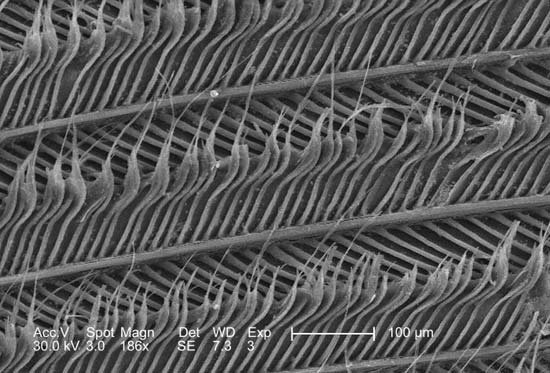
Keratin
Scanning electron micrograph showing strands of keratin in a feather, magnified 186×. |
|
|
| |
|
The weakness of the above classification lies in the fact that many, if not all, globulins contain small amounts of carbohydrate; thus there is no sharp borderline between globulins and mucoproteins. Moreover, the phosphoproteins do not have a prosthetic group that can be isolated; they are merely proteins in which some of the hydroxyl groups of serine are phosphorylated (i.e., contain phosphate). Finally, the globulins include proteins with quite different roles — enzymes, antibodies, fibrous proteins, and contractile proteins. |

|
|
|
|
Classification by biological functions
Classification by biological functions (B)
In view of the unsatisfactory state of the old classification, it is preferable to classify the proteins according to their biological function. Such a classification is far from ideal, however, because one protein can have more than one function. The contractile protein myosin, for example, also acts as an ATPase (adenosine triphosphatase), an enzyme that hydrolyzes adenosine triphosphate (removes a phosphate group from ATP by introducing a water molecule). Another problem with functional classification is that the definite function of a protein frequently is not known. A protein cannot be called an enzyme as long as its substrate (the specific compound upon which it acts) is not known. It cannot even be tested for its enzymatic action when its substrate is not known. |
|
|
|
| |
Special Structure And Function Of Proteins
|
Special Structure And Function Of Proteins
Special Structure And Function Of Proteins (B)
Despite its weaknesses, a functional classification is used here in order to demonstrate, whenever possible, the correlation between the structure and function of a protein. The structural, fibrous proteins are presented first, because their structure is simpler than that of the globular proteins and more clearly related to their function, which is the maintenance of either a rigid or a flexible structure. |
|
|
|
| |
Structural proteins
|
Scleroproteins
Scleroproteins (B)
|
|
| |
|
| Randomly oriented collagenous fibres of varying size in a thin spread of loose areolar connective tissue (magnified about 370 ×). |
|
| |
|
Collagen is the structural protein of bones, tendons, ligaments, and skin. For many years collagen was considered to be insoluble in water. Part of the collagen of calf skin, however, can be extracted with citrate buffer at pH 3.7. A precursor of collagen called procollagen is converted in the body into collagen. Procollagen has a molecular weight of 120,000. Cleavage of one or a few peptide bonds of procollagen yields collagen, which has three subunits, each with a molecular weight of 95,000; therefore, the molecular weight of collagen is 285,000 (3 × 95,000). The three subunits are wound as spirals around an elongated straight axis. The length of each subunit is 2,900 angstroms, and its diameter is approximately 15 angstroms. The three chains are staggered, so that the trimer has no definite terminal limits.
Collagen differs from all other proteins in its high content of proline and hydroxyproline. Hydroxyproline does not occur in significant amounts in any other protein except elastin. Most of the proline in collagen is present in the sequence glycine–proline-X, in which X is frequently alanine or hydroxyproline. Collagen does not contain cystine or tryptophan and therefore cannot substitute for other proteins in the diet. The presence of proline causes kinks in the peptide chain and thus reduces the length of the amino acid unit from 3.7 angstroms in the extended chain of the β-structure to 2.86 angstroms in the collagen chain. In the intertwined triple helix, the glycines are inside, close to the axis; the prolines are outside.
Native collagen resists the action of trypsin but is hydrolyzed by the bacterial enzyme collagenase. When collagen is boiled with water, the triple helix is destroyed, and the subunits are partially hydrolyzed; the product is gelatin. The unfolded peptide chains of gelatin trap large amounts of water, resulting in a hydrated molecule.
When collagen is treated with tannic acid or with chromium salts, cross links form between the collagen fibres, and it becomes insoluble; the conversion of hide into leather is based on this tanning process. The tanned material is insoluble in hot water and cannot be converted to gelatin. On exposure to water at 62° to 63° C (144° to 145° F), however, the cross links formed by the tanning agents collapse, and the leather contracts irreversibly to about one-third its original volume.
Collagen seems to undergo an aging process in living organisms that may be caused by the formation of cross links between collagen fibres. They are formed by the conversion of some lysine side chains to aldehydes (compounds with the general structure RCHO), and the combination of the aldehydes with the ε-amino groups of intact lysine side chains. The protein elastin, which occurs in the elastic fibres of connective tissue, contains similar cross links and may result from the combination of collagen fibres with other proteins. When cross-linked collagen or elastin is degraded, products of the cross-linked lysine fragments, called desmosins and isodesmosins, are formed.
Keratin
Keratin, the structural protein of epithelial cells in the outermost layers of the skin, has been isolated from hair, nails, hoofs, and feathers. Keratin is completely insoluble in cold or hot water; it is not attacked by proteolytic enzymes (i.e., enzymes that break apart, or lyse, protein molecules), and therefore cannot replace proteins in the diet. The great stability of keratin results from the numerous disulfide bonds of cystine. The amino acid composition of keratin differs from that of collagen. Cystine may account for 24 percent of the total amino acids. The peptide chains of keratin are arranged in approximately equal amounts of antiparallel and parallel pleated sheets, in which the peptide chains are linked to each other by hydrogen bonds between the carbonyl and imino groups.
Reduction of the disulfide bonds to sulfhydryl groups results in dissociation of the peptide chains, the molecular weight of which is 25,000 to 28,000 each. The formation of permanent waves in the beauty treatment of hair is based on partial reduction of the disulfide bonds of hair keratin by thioglycol, or some other mild reducing agent, and subsequent oxidation of the sulfhydryl groups (―SH) in the reoriented hair to disulfide bonds (―S―S―) by exposure to the oxygen of the air.
The length of keratin fibres depends on their water content. They can bind approximately 16 percent of water; this hydration is accompanied by an increase in the length of the fibres of 10 to 12 percent.
The most thoroughly investigated keratin is hair keratin, particularly that of wool. It consists of a mixture of peptides with high and low cystine content. When wool is heated in water to about 90° C (190° F), it shrinks irreversibly. This is attributed to the breakage of hydrogen bonds and other noncovalent bonds; disulfide bonds do not seem to be affected.
Others
The most thoroughly investigated scleroprotein has been fibroin, the insoluble material of silk. The raw silk comprising the cocoon of the silkworm consists of two proteins. One, sericin, is soluble in hot water; the other, fibroin, is not. The amino acid composition of the latter differs from that of all other proteins. It contains large amounts of glycine, alanine, tyrosine, and serine; small amounts of the other amino acids; and no sulfur-containing ones. The peptide chains are arranged in antiparallel β-structures. Fibroin is partly soluble in concentrated solutions of lithium thiocyanate or in mixtures of cupric salts and ethylene diamine. Such solutions contain a protein of molecular weight 170,000, which is a dimer of two subunits.
Little is known about either the scleroproteins of the marine sponges or the insoluble proteins of the cellular membranes of animal cells. Some of the membranes are soluble in detergents; others, however, are detergent-insoluble. |

|
|
|
|
The muscle proteins
The muscle proteins (B)
The total amount of muscle proteins in mammals, including humans, exceeds that of any other protein. About 40 percent of the body weight of a healthy human adult weighing about 70 kilograms (150 pounds) is muscle, which is composed of about 20 percent muscle protein. Thus, the human body contains about 5 to 6 kilograms (11 to 13 pounds) of muscle protein. An albumin-like fraction of these proteins, originally called myogen, contains various enzymes—phosphorylase, aldolase, glyceraldehyde phosphate dehydrogenase, and others; it does not seem to be involved in contraction. The globulin fraction contains myosin, the contractile protein, which also occurs in blood platelets, small bodies found in blood. Similar contractile substances occur in other contractile structures; for example, in the cilia or flagella (whiplike organs of locomotion) of bacteria and protozoans. In contrast to the scleroproteins, the contractile proteins are soluble in salt solutions and susceptible to enzymatic digestion.
The energy required for muscle contraction is provided by the oxidation of carbohydrates or lipids. The term mechanochemical reaction has been used for this conversion of chemical into mechanical energy. The molecular process underlying the reaction is known to involve the fibrous muscle proteins, the peptide chains of which undergo a change in conformation during contraction.
Myosin, which can be removed from fresh muscle by adding it to a chilled solution of dilute potassium chloride and sodium bicarbonate, is insoluble in water. Myosin, solutions of which are highly viscous, consists of an elongated—probably double-stranded—peptide chain, which is coiled at both ends in such a way that a terminal globule is formed. The length of the molecule is approximately 160 nanometres and its average diameter 2.6 nanometres. The equivalent weight of each of the two terminal globules is approximately 30,000; the molecular weight of myosin is close to 500,000. Trypsin splits myosin into large fragments called meromyosin. Myosin contains many amino acids with positively and negatively charged side chains; they form 18 and 16 percent, respectively, of the total number of amino acids. Myosin catalyzes the hydrolytic cleavage of ATP (adenosine triphosphate). A smaller protein with properties similar to those of myosin is tropomyosin. It has a molecular weight of 70,000 and dimensions of 45 by 2 nanometres. More than 90 percent of its peptide chains are present in the α-helix form.
Myosin combines easily with another muscle protein called actin, the molecular weight of which is about 50,000; it forms 12 to 15 percent of the muscle proteins. Actin can exist in two forms—one, G-actin, is globular; the other, F-actin, is fibrous. Actomyosin is a complex molecule formed by one molecule of myosin and one or two molecules of actin. In muscle, actin and myosin filaments are oriented parallel to each other and to the long axis of the muscle. The actin filaments are linked to each other lengthwise by fine threads called S filaments. During contraction the S filaments shorten, so that the actin filaments slide toward each other, past the myosin filaments, thus causing a shortening of the muscle (for a detailed description of the process, see muscle: Striated muscle). |
| |
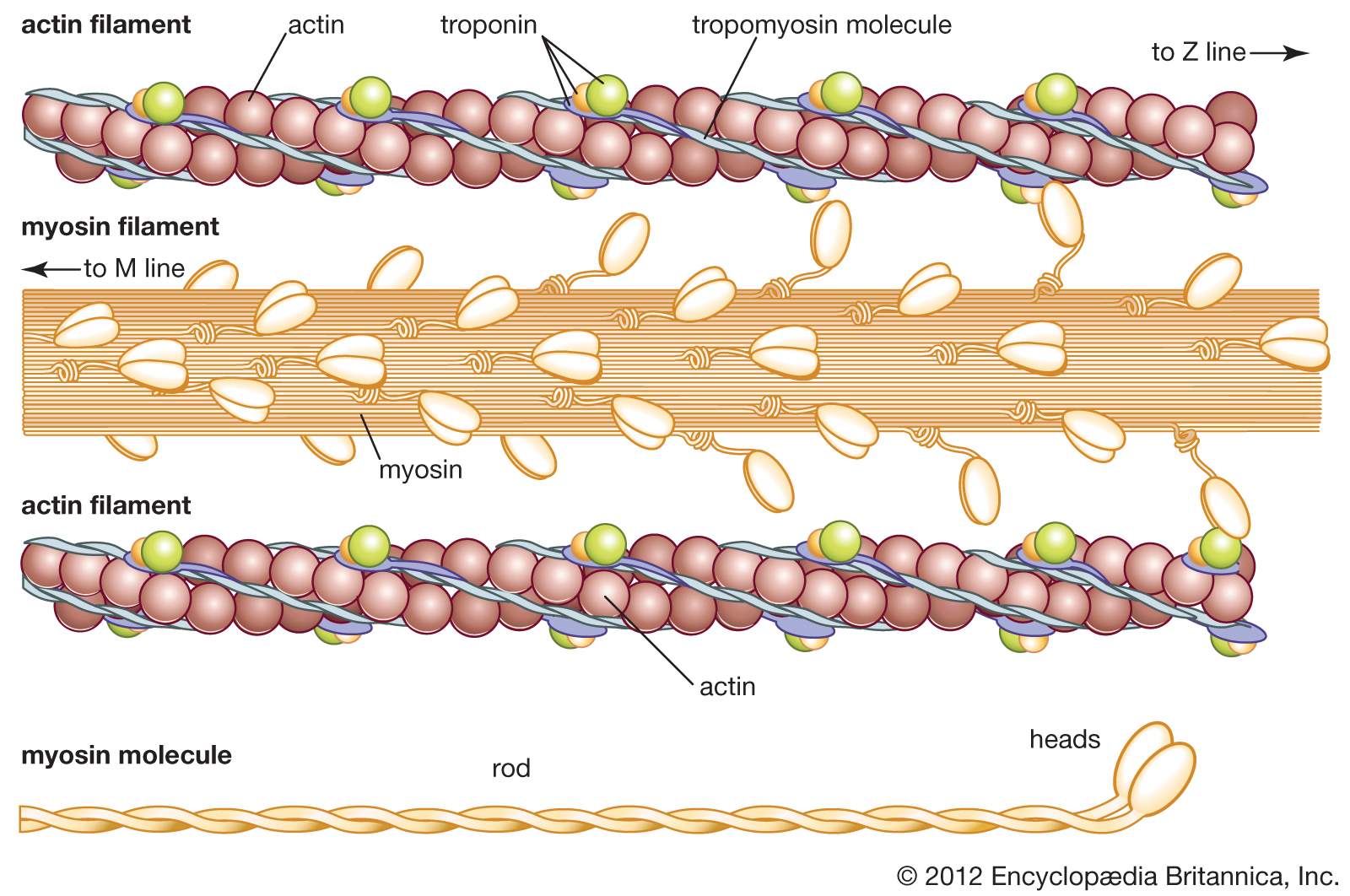
Muscle: actin and myosin
The structure of actin and myosin filaments. |
|

|
|
|
|
Fibrinogen and fibrin
Fibrinogen and fibrin (B)
Fibrinogen, the protein of the blood plasma, is converted into the insoluble protein fibrin during the clotting process. The fibrinogen-free fluid obtained after removal of the clot, called blood serum, is blood plasma minus fibrinogen. The fibrinogen content of the blood plasma is 0.2 to 0.4 percent. |
| |
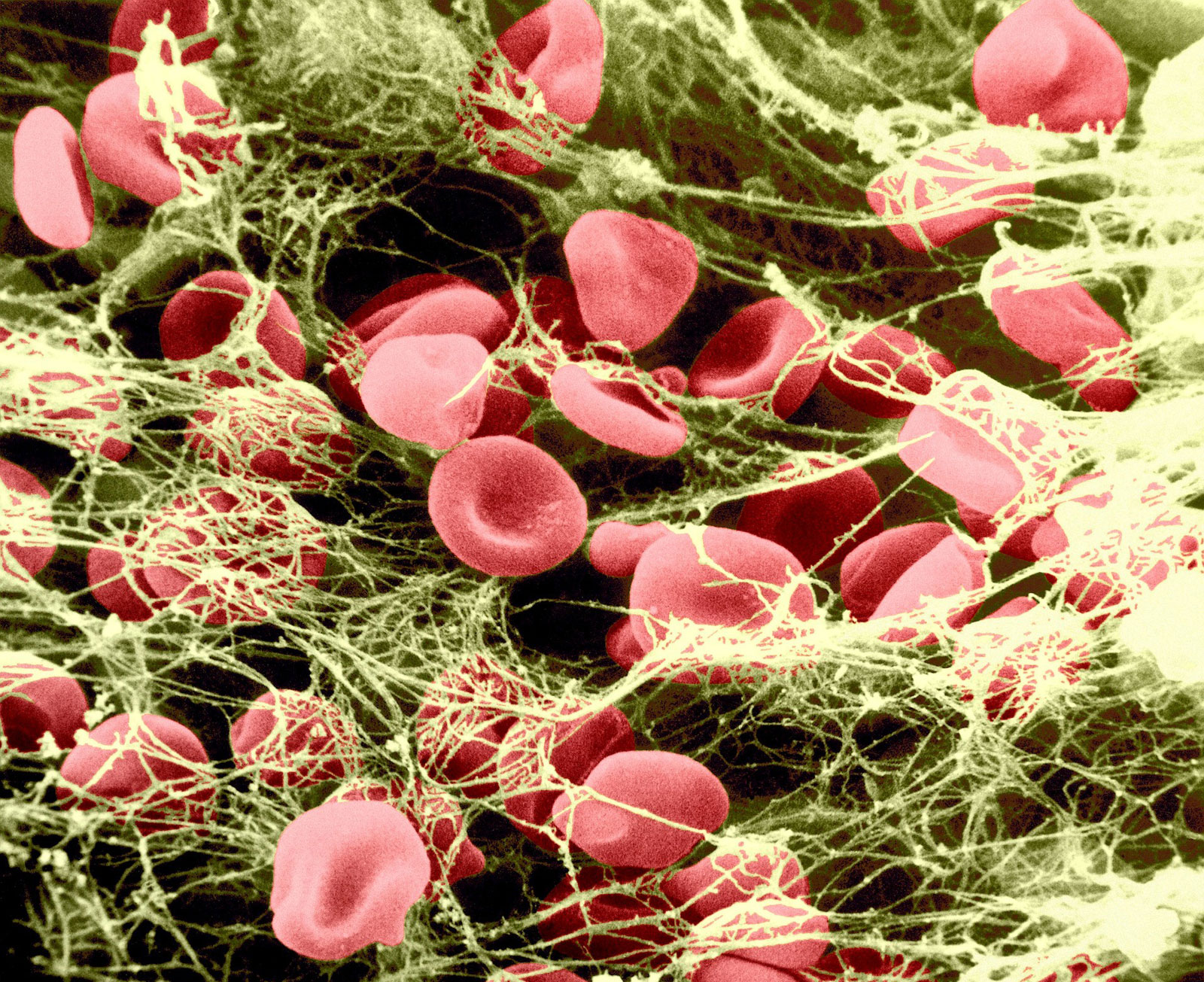
Fibrin in blood clotting
Red blood cells (erythrocytes) trapped in a mesh of fibrin threads. Fibrin, a tough, insoluble protein formed after injury to the blood vessels, is an essential component of blood clots. |
| |
|
Fibrinogen can be precipitated from the blood plasma by half-saturation with sodium chloride. Fibrinogen solutions are highly viscous and show strong flow birefringence. In electron micrographs the molecules appear as rods with a length of 47.5 nanometres and a diameter of 1.5 nanometres; in addition, two terminal and a central nodule are visible. The molecular weight is 340,000. An unusually high percentage, about 36 percent, of the amino acid side chains are positively or negatively charged.
The clotting process is initiated by the enzyme thrombin, which catalyzes the breakage of a few peptide bonds of fibrinogen; as a result, two small fibrinopeptides with molecular weights of 1,900 and 2,400 are released. The remainder of the fibrinogen molecule, a monomer, is soluble and stable at pH values less than 6 (i.e., in acid solutions). In neutral solution (pH 7) the monomer is converted into a larger molecule, insoluble fibrin; this results from the formation of new peptide bonds. The newly formed peptide bonds form intermolecular and intramolecular cross links, thus giving rise to a large clot, in which all molecules are linked to each other. Clotting, which takes place only in the presence of calcium ions, can be prevented by compounds such as oxalate or citrate, which have a high affinity for calcium ions. |

|
|
|
|
Albumins, globulins, and other soluble proteins
Albumins, globulins, and other soluble proteins (B)
The blood plasma, the lymph, and other animal fluids usually contain one to seven grams of protein per 100 millilitres of fluid, which includes small amounts of hundreds of enzymes and a large number of protein hormones. The discussion below is limited largely to the proteins that occur in large amounts and can be easily isolated from the body fluids. |
|
|
|
Proteins of the blood serum
Proteins of the blood serum (B)
Human blood serum contains about 7 percent protein, two-thirds of which is in the albumin fraction; the other third is in the globulin fraction. Electrophoresis of serum reveals a large albumin peak and three smaller globulin peaks, the alpha-, beta-, and gamma-globulins. The amounts of alpha-, beta-, and gamma-globulin in normal human serum are approximately 1.5, 1.9, and 1.1 percent, respectively. Each globulin fraction is a mixture of many different proteins, as has been demonstrated by immunoelectrophoresis. In this method, serum from an animal (e.g., a rabbit) injected with human serum is allowed to diffuse into the four protein bands—albumin, alpha-, beta-, and gamma-globulin—obtained from the electrophoresis of human serum. Because the animal has previously been injected with human serum, its blood contains antibodies (substances formed in response to a foreign substance introduced into the body) against each of the human serum proteins; each antibody combines with the serum protein (antigen) that caused its formation in the animal. The result is the formation of about 20 regions of insoluble antigen-antibody precipitate, which appear as white arcs in the transparent gel of the electrophoresis medium. Each region corresponds to a different human serum protein.
Serum albumin is much less heterogeneous (i.e., contains fewer distinct proteins) than are the globulins; in fact, it is one of the few serum proteins that can be obtained in a crystalline form. Serum albumin combines easily with many acidic dyes (e.g., Congo red and methyl orange); with bilirubin, the yellow bile pigment; and with fatty acids. It seems to act, in living organisms, as a carrier for certain biological substances. Present in blood serum in relatively high concentration, serum albumin also acts as a protective colloid, a protein that stabilizes other proteins. Albumin (molecular weight of 68,000) has a single free sulfhydryl (―SH) group, which on oxidation forms a disulfide bond with the sulfhydryl group of another serum albumin molecule, thus forming a dimer. The isoelectric point of serum albumin is pH 4.7.
The alpha-globulin fraction of blood serum is a mixture of several conjugated proteins. The best known are an α-lipoprotein (combination of lipid and protein) and two mucoproteins (combinations of carbohydrate and protein). One mucoprotein is called orosomucoid, or α1-acid glycoprotein; the other is called haptoglobin because it combines specifically with globin, the protein component of hemoglobin. Haptoglobin contains about 20 percent carbohydrate. The beta-globulin fraction of serum contains, in addition to lipoproteins and mucoproteins, two metal-binding proteins, transferrin and ceruloplasmin, which bind iron and copper, respectively. They are the principal iron and copper carriers of the blood.
The gamma-globulins are the most heterogeneous globulins. Although most have a molecular weight of approximately 150,000, that of some, called macroglobulins, is as high as 800,000. Because typical antibodies are of the same size and exhibit the same electrophoretic behaviour as γ-globulins, they are called immunoglobulins. The designation IgM or gamma M (γM) is used for the macroglobulins; the designation IgG or gamma G (γG) is used for γ−globulins of molecular weight 150,000. |
| |
Antibody structure
The four-chain structure of an antibody, or immunoglobulin, molecule. The basic unit is composed of two identical light (L) chains and two identical heavy (H) chains, which are held together by disulfide bonds to form a flexible Y shape. Each chain is composed of a variable (V) region and a constant (C) region. |
|

|
|
|
|
Milk proteins
Milk proteins (B)
Milk contains the following: an albumin, α-lactalbumin; a globulin, beta-lactoglobulin; and a phosphoprotein, casein. If acid is added to milk, casein precipitates. The remaining watery liquid (the supernatant solution), or whey, contains α-lactalbumin and β-lactoglobulin. Both have been obtained in crystalline form; in bovine milk, their molecular weights are approximately 14,000 and 18,400, respectively. Lactoglobulin also occurs as a dimer of molecular weight 37,000. Genetic variations can produce small variations in the amino acid composition of lactoglobulin. The amino acid composition and the tertiary structure of lactalbumin resemble that of lysozyme, an egg protein.
Casein is precipitated not only by the addition of acid but also by the action of the enzyme rennin, which is found in gastric juice. Rennin from calf stomachs is used to precipitate casein, from which cheese is made. Milk fat precipitates with casein; milk sugar, however, remains in the supernatant (whey). Casein is a mixture of several similar phosphoproteins, called α-, β-, γ−, and κ-casein, all of which contain some serine side chains combined with phosphoric acid. Approximately 75 percent of casein is α-casein. Cystine has been found only in κ-casein. In milk, casein seems to form polymeric globules (micelles) with radially arranged monomers, each with a molecular weight of 24,000; the acidic side chains occur predominantly on the surface of the micelle, rather than inside. |

|
|
|
|
Egg proteins
Egg proteins (B)
About 50 percent of the proteins of egg white are composed of ovalbumin, which is easily obtained in crystals. Its molecular weight is 46,000 and its amino acid composition differs from that of serum albumin. Other proteins of egg white are conalbumin, lysozyme, ovoglobulin, ovomucoid, and avidin. Lysozyme is an enzyme that hydrolyzes the carbohydrates found in the capsules certain bacteria secrete around themselves; it causes lysis (disintegration) of the bacteria. The molecular weight of lysozyme is 14,100. Its three-dimensional structure is similar to that of α-lactalbumin, which stimulates the formation of lactose by the enzyme lactose synthetase. Lysozyme has also been found in the urine of patients suffering from leukemia, meningitis, and renal disease.
Avidin is a glycoprotein that combines specifically with biotin, a vitamin. In animals fed large amounts of raw egg white, the action of avidin results in “egg-white injury.” The molecular weight of avidin, which forms a tetramer, is 16,200. Its amino acid sequence is known.
Egg-yolk proteins contain a mixture of lipoproteins and livetins. The latter are similar to serum albumin, α-globulin, and β-globulin. The yolk also contains a phosphoprotein, phosvitin. Phosvitin, which has also been found in fish sperm, has a molecular weight of 40,000 and an unusual amino acid composition; one third of its amino acids are phosphoserine. |

|
|
|
|
Protamines and histones
Protamines and histones (B)
Protamines are found in the sperm cells of fish. The most thoroughly investigated protamines are salmine from salmon sperm and clupeine from herring sperm. The protamines are bound to deoxyribonucleic acid (DNA), forming nucleoprotamines. The amino acid composition of the protamines is simple; they contain, in addition to large amounts of arginine, small amounts of five or six other amino acids. The composition of the salmine molecule, for example, is: Arg51, Ala4, Val4, Ile1, Pro7, and Ser6, in which the subscript numbers indicate the number of each amino acid in the molecule. Because of the high arginine content, the isoelectric points of the protamines are at pH values of 11 to 12; i.e., the protamines are alkaline. The molecular weights of salmine and clupeine are close to 6,000. All of the protamines investigated thus far are mixtures of several similar proteins.
The histones are less basic than the protamines. They contain high amounts of either lysine or arginine and small amounts of aspartic acid and glutamic acid. Histones occur in combination with DNA as nucleohistones in the nuclei of the body cells of animals and plants, but not in animal sperm. The molecular weights of histones vary from 10,000 to 22,000. In contrast to the protamines, the histones contain most of the 20 amino acids, with the exception of tryptophan and the sulfur-containing ones. Like the protamines, histone preparations are heterogeneous mixtures. The amino acid sequence of some of the histones has been determined. |

|
|
|
|
Plant proteins
Plant proteins (B)
Plant proteins, mostly globulins, have been obtained chiefly from the protein-rich seeds of cereals and legumes. Small amounts of albumins are found in seeds. The best known globulins, insoluble in water, can be extracted from seeds by treatment with 2 to 10 percent solutions of sodium chloride. Many plant globulins have been obtained in crystalline form; they include edestin from hemp, molecular weight 310,000; amandin from almonds, 330,000; concanavalin A (42,000) and B (96,000); and canavalin (113,000) from jack beans. They are polymers of smaller subunits; edestin, for example, is a hexamer of a subunit with a molecular weight of 50,000, and concanavalin B a trimer of a subunit with a molecular weight of 30,000. After extraction of lipids from cereal seeds by ether and alcohol, further extraction with water containing 50 to 80 percent of alcohol yields proteins that are insoluble in water but soluble in water–ethanol mixtures and have been called prolamins. Their solubility in aqueous ethanol may result from their high proline and glutamine content. Gliadin, the prolamin from wheat, contains 14 grams of proline and 46 grams of glutamic acid in 100 grams of protein; most of the glutamic acid is in the form of glutamine. The total amounts of the basic amino acids (arginine, lysine, and histidine) in gliadin are only 5 percent of the weight of gliadin. Because the glysine content is either low or nonexistent, human populations dependent on grain as a sole protein source suffer from lysine deficiency. |

|
|
|
|
| |
Conjugated proteins
|
Combination of proteins with prosthetic groups
Combination of proteins with prosthetic groups (B)
The link between a protein molecule and its prosthetic group is a covalent bond (an electron-sharing bond) in the glycoproteins, the biliproteins, and some of the heme proteins. In lipoproteins, nucleoproteins, and some heme proteins, the two components are linked by noncovalent bonds; the bonding results from the same forces that are responsible for the tertiary structure of proteins: hydrogen bonds, salt bridges between positively and negatively charged groups, disulfide bonds, and mutual interaction of hydrophobic groups. In the metalloproteins (proteins with a metal element as a prosthetic group), the metal ion usually forms a centre to which various groups are bound.
Some of the conjugated proteins have been mentioned in preceding sections because they occur in the blood serum, in milk, and in eggs; others are discussed below in sections dealing with respiratory proteins and enzymes.
Mucoproteins and glycoproteins
The prosthetic groups in mucoproteins and glycoproteins are oligosaccharides (carbohydrates consisting of a small number of simple sugar molecules) usually containing from four to 12 sugar molecules; the most common sugars are galactose, mannose, glucosamine, and galactosamine. Xylose, fucose, glucuronic acid, sialic acid, and other simple sugars sometimes also occur. Some mucoproteins contain 20 percent or more of carbohydrate, usually in several oligosaccharides attached to different parts of the peptide chain. The designation mucoprotein is used for proteins with more than 3 to 4 percent carbohydrate; if the carbohydrate content is less than 3 percent, the protein is sometimes called a glycoprotein or simply a protein.
Mucoproteins, highly viscous proteins originally called mucins, are found in saliva, in gastric juice, and in other animal secretions. Mucoproteins occur in large amounts in cartilage, synovial fluid (the lubricating fluid of joints and tendons), and egg white. The mucoprotein of cartilage is formed by the combination of collagen with chondroitinsulfuric acid, which is a polymer of either glucuronic or iduronic acid and acetylhexosamine or acetylgalactosamine. It is not yet clear whether or not chondroitinsulfate is bound to collagen by covalent bonds.
The bond between the protein and the lipid portion of lipoproteins and proteolipids is a noncovalent one. It is thought that some of the lipid is enclosed in a meshlike arrangement of peptide chains and becomes accessible for reaction only after the unfolding of the chains by denaturing agents. Although lipoproteins in the α- and β-globulin fraction of blood serum are soluble in water (but insoluble in organic solvents), some of the brain lipoproteins, because they have a high lipid content, are soluble in organic solvents; they are called proteolipids. The β-lipoprotein of human blood serum is a macroglobulin with a molecular weight of about 1,300,000, 70 percent of which is lipid; of the lipid, about 30 percent is phospholipid and 40 percent cholesterol and compounds derived from it. Because of their lipid content, the lipoproteins have the lowest density (mass per unit volume) of all proteins and are usually classified as low- and high-density lipoproteins (LDL and HDL).
Coloured lipoproteins are formed by the combination of protein with carotenoids. Crustacyanin, the pigment of lobsters, crayfish, and other crustaceans, contains astaxanthin, which is a compound derived from carotene. Among the most interesting of the coloured lipoproteins are the pigments of the retina of the eye. They contain retinal, which is a compound derived from carotene and which is formed by the oxidation of vitamin A. In rhodopsin, the red pigment of the retina, the aldehyde group (―CHO) of retinal forms a covalent bond with an amino (―NH2) group of opsin, the protein carrier. Colour vision is mediated by the presence of several visual pigments in the retina that differ from rhodopsin either in the structure of retinal or in that of the protein carrier.
Metalloproteins
Proteins in which heavy metal ions are bound directly to some of the side chains of histidine, cysteine, or some other amino acid are called metalloproteins. Two metalloproteins, transferrin and ceruloplasmin, occur in the globulin fractions of blood serum; they act as carriers of iron and copper, respectively. Transferrin has a molecular weight of about 80,000 and consists of two identical subunits, each of which contains one ferric ion (Fe3+) that seems to be bound to tyrosine. Several genetic variants of transferrin are known to occur in humans. Another iron protein, ferritin, which contains 20 to 22 percent iron, is the form in which iron is stored in animals; it has been obtained in crystalline form from liver and spleen. A molecule consisting of 20 subunits, its molecular weight is approximately 480,000. The iron can be removed by reduction from the ferric (Fe3+) to the ferrous (Fe2+) state. The iron-free protein, apoferritin, is synthesized in the body before the iron is incorporated.
Green plants and some photosynthetic and nitrogen-fixing bacteria (i.e., bacteria that convert atmospheric nitrogen, N2, into amino acids and proteins) contain various ferredoxins. They are small proteins containing 50 to 100 amino acids and a chain of iron and disulfide units (FeS2), in which some of the sulfur atoms are contributed by cysteine; others are sulfide ions (S2−). The number of FeS2 units per ferredoxin molecule varies from five in the ferredoxin of spinach to 10 in the ferredoxin of certain bacteria. Ferredoxins act as electron carriers in photosynthesis and in nitrogen fixation.
Ceruloplasmin is a copper-containing globulin that has a molecular weight of 151,000; the molecule consists of eight subunits, each containing one copper ion. Ceruloplasmin is the principal carrier of copper in organisms, although copper can also be transported by the iron-containing globulin transferrin. Another copper-containing protein, copper-zinc superoxide dismutase (formerly known as erythrocuprein), has been isolated from red blood cells; it has also been found in the liver and in the brain. The molecule, which consists of two subunits of similar size, contains copper ions and zinc ions. Because of their copper content, ceruloplasmin and copper-zinc superoxide dismutase possess catalytic activity in oxidation-reduction reactions.
Many animal enzymes contain zinc ions, which are usually bound to the sulfur of cysteine. Horse kidneys contain the protein metallothionein, which contain zinc and cadmium; both are bound to sulfur. A vanadium-protein complex (hemovanadin) has been found in surprisingly high amounts in yellowish-green cells (vanadocytes) of tunicates, which are marine invertebrates.
Heme proteins and other chromoproteins
Although the heme proteins contain iron, they are usually not classified as metalloproteins, because their prosthetic group is an iron- porphyrin complex in which the iron is bound very firmly. The intense red or brown colour of the heme proteins is not caused by iron but by porphyrin, a complex cyclic structure. All porphyrin compounds absorb light intensely at or close to 410 nanometres. Porphyrin consists of four pyrrole rings (five-membered closed structures containing one nitrogen and four carbon atoms) linked to each other by methine groups (―CH=). The iron atom is kept in the centre of the porphyrin ring by interaction with the four nitrogen atoms. The iron atom can combine with two other substituents; in oxyhemoglobin, one substituent is a histidine of the protein carrier, the other is an oxygen molecule. In some heme proteins, the protein is also bound covalently to the side chains of porphyrin. Heme proteins are described below (see Respiratory proteins).
The chromoprotein melanin, a pigment found in dark skin, dark hair, and melanotic tumours, occurs in every major group of living organisms and appears to be remarkably diverse in structure. In humans, melanin produced by melanocytes may be dark brown (eumelanin) or pale red or yellowish (phaeomelanin). The different types are synthesized via different pathways, though they share the same initial step—the oxidation of tyrosine.
Green chromoproteins called biliproteins are found in many insects, such as grasshoppers, and also in the eggshells of many birds. The biliproteins are derived from the bile pigment biliverdin, which in turn is formed from porphyrin; biliverdin contains four pyrrole rings and three of the four methine groups of porphyrin. Large amounts of biliproteins have been found in red algae and blue-green algae; the red protein is called phycoerythrin, the blue one phycocyanobilin.
Nucleoproteins
When a protein solution is mixed with a solution of a nucleic acid, the phosphoric acid component of the nucleic acid combines with the positively charged ammonium groups (―NH3+) of the protein to form a protein–nucleic acid complex. The nucleus of a cell contains predominantly deoxyribonucleic acid (DNA) and the cytoplasm predominantly ribonucleic acid (RNA); both parts of the cell also contain protein. Protein–nucleic acid complexes, therefore, form in living cells.
The only nucleoproteins for which some evidence for specificity exists are nucleoprotamines, nucleohistones, and some RNA and DNA viruses. The nucleoprotamines are the form in which protamines occur in the sperm cells of fish; the histones of the thymus and of pea seedlings and other plant material apparently occur predominantly as nucleohistones. Both nucleoprotamines and nucleohistones contain only DNA.
Some of the simplest viruses consist of a specific RNA, which is coated by protein. One of the best known RNA viruses, tobacco mosaic virus (TMV), has the shape of a rod. RNA comprises only 5.1 percent of the mass of the virus. The complete sequence of the virus protein, which consists of about 2,130 identical peptide chains, each containing 158 amino acids, has been determined. The protein is arranged in a spiral around the RNA core. |
| |

Tobacco mosaic virus
Schematic structure of the tobacco mosaic virus. The cutaway section shows the helical ribonucleic acid associated with protein molecules in a ratio of three nucleotides per protein molecule.
|
| |
| DNA has been found in most bacterial viruses (bacteriophages) and in some animal viruses. As in TMV, the core of DNA is surrounded by protein. Phage protein is a mixture of enzymes and therefore cannot be considered as the protein portion of only one nucleoprotein. |
|
| |

|
|
|
|
Respiratory proteins
Respiratory proteins (B)
Hemoglobin is the oxygen carrier in all vertebrates and some invertebrates. In oxyhemoglobin (HbO2), which is bright red, the ferrous ion (Fe2+) is bound to the four nitrogen atoms of porphyrin; the other two substituents are an oxygen molecule and the histidine of globin, the protein component of hemoglobin. Deoxyhemoglobin (deoxy-Hb), as its name implies, is oxyhemoglobin minus oxygen (i.e., reduced hemoglobin); it is purple in colour. Oxidation of the ferrous ion of hemoglobin yields a ferric compound, methemoglobin, sometimes called hemiglobin or ferrihemoglobin. The oxygen of oxyhemoglobin can be displaced by carbon monoxide, for which hemoglobin has a much greater affinity, preventing oxygen from reaching the body tissues.
The hemoglobins of all mammals, birds, and many other vertebrates are tetramers of two α- and two β-chains. The molecular weight of the tetramer is 64,500; the molecular weight of the α- and β-chains is approximately 16,100 each, and the four subunits are linked to each other by noncovalent interactions. If hemin (the ferric porphyrin component) is removed from globin (the protein component), two molecules of globin, each consisting of one α- and one β-chain, are obtained; the molecular weight of globin is 32,200. In contrast to hemoglobin, globin is an unstable protein that is easily denatured. If native globin is incubated with a solution of hemin at pH values of 8 to 9, native hemoglobin is reconstituted. Myoglobin, the red pigment of mammalian muscles, is a monomer with a molecular weight of 16,000.
The mammalian hemoglobins differ from each other in their amino acid composition and therefore in their secondary and tertiary structure. Rat and horse hemoglobins crystallize very easily, but those of humans, cattle, and sheep, because they are more soluble, are difficult to crystallize. The shape of hemoglobin crystals varies in different species; moreover, decomposition and denaturation occur at different rates in different species. It was also found that the blood of human newborns contains two different hemoglobins: about 20 percent of their hemoglobin is an adult hemoglobin (hemoglobin A) and 80 percent is a fetal hemoglobin (hemoglobin F). Hemoglobin F persists in the infant for the first seven months of life. The same hemoglobin F has also been found in the blood of patients suffering from thalassemia, an anemia with a high incidence in regions surrounding the Mediterranean Sea. Hemoglobin F contains, as does hemoglobin A, two α-chains; the two β-chains, however, have been replaced by two quite different γ-chains. When the technique of electrophoresis was first applied to the hemoglobin of blacks suffering from sickle cell anemia in 1949, a new hemoglobin (hemoglobin S) was discovered. More than 200 different human hemoglobins have been discovered since. They differ from normal hemoglobin A in the amino acid composition of either the α- or the β-chain.
The hemoglobins of some of the lowest fishes are monomers containing one iron atom per molecule. Hemoglobin-like respiratory proteins have been found in some invertebrates. The red hemoglobin of insects, mollusks, and protozoans is called erythrocruorin. It differs from vertebrate hemoglobin by its high molecular weight.
Although green plants contain no hemoglobin, a red protein, called leghemoglobin, has been discovered in the root nodules of leguminous plants. It seems to be produced by the nitrogen-fixing bacteria of the root nodules and may be involved in the reduction of atmospheric nitrogen to ammonia and amino acids.
Other respiratory proteins
A green respiratory protein, chlorocruorin, has been found in the blood of marine worms in the genera Serpula and Spirographis. It has the same high molecular weight as erythrocruorin but differs from hemoglobin in its prosthetic group. A red metalloprotein, hemerythrin, acts as a respiratory protein in marine worms of the phylum Sipuncula. The molecule consists of eight subunits with a molecular weight of 13,500 each. Hemerythrin contains no porphyrins and therefore is not a heme protein.
A metalloprotein containing copper is the respiratory protein of crustaceans (shrimps, crabs, etc.) and of some gastropods (snails). The protein, called hemocyanin, is pale yellow when not combined with oxygen, and blue when combined with oxygen. The molecular weights of hemocyanins vary from 300,000 to 9,000,000. Each animal investigated thus far apparently has a species-specific hemocyanin. |

|
|
|
|
| |
Protein hormones
|
Protein hormones
Protein hormones (B)
Some hormones that are products of endocrine glands are proteins or peptides, others are steroids. (The origin of hormones, their physiological role, and their mode of action are dealt with in the article hormone.) None of the hormones has any enzymatic activity. Each has a target organ in which it elicits some biological action — e.g., secretion of gastric or pancreatic juice, production of milk, production of steroid hormones. The mechanism by which the hormones exert their effects is not fully understood. Cyclic adenosine monophosphate is involved in the transmittance of the hormonal stimulus to the cells whose activity is specifically increased by the hormone. |
|
|
|
Hormones of the thyroid gland
Hormones of the thyroid gland (B)
Thyroglobulin, the active groups of which are two molecules of the iodine-containing compound thyroxine, has a molecular weight of 670,000. Thyroglobulin also contains thyroxine with two and three iodine atoms instead of four and tyrosine with one and two iodine atoms. Injection of the hormone causes an increase in metabolism; lack of it results in a slowdown.
Another hormone, calcitonin, which lowers the calcium level of the blood, occurs in the thyroid gland. The amino acid sequences of calcitonin from pig, beef, and salmon differ from human calcitonin in some amino acids. All of them, however, have the half-cystines (C) and the prolinamide (P) in the same position. |
| |
|
| Parathyroid hormone (parathormone), produced in small glands that are embedded in or lie behind the thyroid gland, is essential for maintaining the calcium level of the blood. A decrease in its production results in hypocalcemia (a reduction of calcium levels in the bloodstream below the normal range). Bovine parathormone has a molecular weight of 8,500; it contains no cystine or cysteine and is rich in aspartic acid, glutamic acid, or their amides. |

|
|
|
|
Hormones of the pancreas
Hormones of the pancreas (B)
Although the amino acid structure of insulin has been known since 1949, repeated attempts to synthesize it gave very poor yields because of the failure of the two peptide chains to combine forming the correct disulfide bridge. The ease of the biosynthesis of insulin is explained by the discovery in the pancreas of proinsulin, from which insulin is formed. The single peptide chain of proinsulin loses a peptide consisting of 33 amino acids and called the connecting peptide, or C peptide, during its conversion to insulin. The disulfide bridges of proinsulin connect the A and B chains. |
| |
|
In aqueous solutions, insulin exists predominantly as a complex of six subunits, each of which contains an A and a B chain. The insulins of several species have been isolated and analyzed; their amino acid sequences have been found to differ somewhat, but all apparently contain the same disulfide bridges between the two chains.
Although the injection of insulin lowers the blood sugar, administration of glucagon, another pancreas hormone, raises the blood sugar level. Glucagon consists of a straight peptide chain of 29 amino acids. It has been synthesized; the synthetic product has the full biological activity of natural glucagon. The structure of glucagon is free of cystine and isoleucine.
The pituitary gland has an anterior lobe, a posterior lobe, and an intermediate portion; they differ in cellular structure and in the structure and action of the hormones they form. The posterior lobe produces two similar hormones, oxytocin and vasopressin. The former causes contraction of the pregnant uterus; the latter raises the blood pressure. Both are octapeptides formed by a ring of five amino acids (the two cystine halves count as one amino acid) and a side chain of three amino acids. The two cystine halves are linked to each other by a disulfide bond, and the C terminal amino acid is glycinamide. The structure has been established and confirmed. Human vasopressin differs from oxytocin in that isoleucine is replaced by phenylalanine and leucine by arginine. |
| |
|
| The intermediate part of the pituitary gland produces the melanocyte-stimulating hormone (MSH), which causes expansion of the pigmented melanophores (cells) in the skin of frogs and other batrachians. Two hormones, called α-MSH and β-MSH, have been prepared from hog pituitary glands. The first, α-MSH, consists of 13 amino acids; its N terminal serine is acetylated (i.e., the acetyl group, CH3CO, of acetic acid is attached), and its C terminal valine residue is present as valinamide. The second, β-MSH, contains in its 18 amino acids many of those occurring in α-MSH. |
| |
|
| The anterior pituitary lobe produces several protein hormones—a thyroid-stimulating hormone (thyrotropin), molecular weight 28,000; a lactogenic hormone, molecular weight 22,500; a growth hormone, molecular weight 21,500; a luteinizing hormone, molecular weight 30,000; and a follicle-stimulating hormone, molecular weight 29,000. The thyroid-stimulating hormone consists of α and β subunits with a composition similar to the subunits of luteinizing hormone. When separated, neither of the two subunits has hormonal activity; when combined, however, they regain about 50 percent of the original activity. The lactogenic hormone (prolactin) from sheep pituitary glands contains 190 amino acids. Their sequence has been elucidated; a similar peptide chain of 188 amino acids that has been synthesized not only has 10 percent of the biological activity of the natural hormone but also some activity of the growth hormone. The amino acid sequence of the growth hormone (somatotropic hormone) is also known; it seems to stimulate the synthesis of RNA and in this way to accelerate growth. The luteinizing hormone, a mucoprotein containing about 12 percent carbohydrate, consists of two subunits, each with a molecular weight of approximately 15,000; when separated, the subunits recombine spontaneously. The urine of pregnant women contains chorionic gonadotropin, the presence of which makes possible early diagnosis of pregnancy. The amino acid sequence is known. The sequence of 160 of its 190 amino acids is identical with those of the growth hormone; 100 of these also occur in the same sequence as in lactogenic hormone. The different pituitary hormones and the chorionic gonadotropin thus may have been derived from a common substance that, during evolution, underwent differentiation. |

|
|
|
|
Peptides with hormonelike activity
Peptides with hormonelike activity (B)
Small peptides have been discovered that, like hormones, act on certain target organs. One peptide, angiotensin (angiotonin or hypertensin), is formed in the blood from angiotensinogen by the action of renin, an enzyme of the kidney. It is an octapeptide and increases blood pressure. Similar peptides include bradykinin, which stimulates smooth muscles; gastrin, which stimulates secretion of hydrochloric acid and pepsin in the stomach; secretin, which stimulates the flow of pancreatic juice; and kallikrein, the activity of which is similar to bradykinin. |
|
|
|
| |
Immunoglobulins and antibodies
|
Immunoglobulins and antibodies
Immunoglobulins and antibodies (B)
Antibodies, proteins that combat foreign substances in the body, are associated with the globulin fraction of the immune serum. As stated previously, when the serum globulins are separated into α-, β-, and γ- fractions, antibodies are associated with the γ-globulins. Antibodies can be purified by precipitation with the antigen (i.e., the foreign substance) that caused their formation, followed by separation of the antigen-antibody complex. Antibodies prepared in this way consist of a mixture of many similar antibody molecules, which differ in molecular weight, amino acid composition, and other properties. The same differences are found in the γ-globulins of normal blood serums. The γ-globulin of normal blood serum is thought to consist of a mixture of hundreds of different γ-globulins, each of which occurs in amounts too small for isolation. Because the physical and chemical properties of normal γ-globulins are the same as those of antibodies, the γ-globulins are frequently called immunoglobulins. They may be considered to be antibodies against unknown antigens. If solutions of γ-globulin are resolved by gel filtration through dextran, the first fraction has a molecular weight of 900,000. This fraction is called IgM or γM; Ig is an abbreviation for immunoglobulin and M for macroglobulin. The next two fractions are IgA (γA) and IgG (γG), with molecular weights of about 320,000 and 150,000 respectively. Two other immunoglobulins, known as IgD and IgE, have also been detected in much smaller amounts in some immune sera.
The bulk of the immunoglobulins is found in the IgG fraction, which also contains most of the antibodies. The IgM molecules are apparently pentamers—aggregates of five of the IgG molecules. Electron microscopy shows their five subunits to be linked to each other by disulfide bonds in the form of a pentagon. The IgA molecules are found principally in milk and in secretions of the intestinal mucosa. Some of them contain, in addition to a dimer of IgG, a “secretory piece” that enables the passage of IgA molecules between tissue and fluid; the structure of the secretory piece is not yet known. The IgM and IgA immunoglobulins and antibodies contain 10 to 15 percent carbohydrate; the carbohydrate content of the IgG molecules is 2 to 3 percent.
IgG molecules treated with the enzyme papain split into three fragments of almost identical molecular weight of 50,000. Two of these, called Fab fragments, are identical; the third is abbreviated Fc. Reduction to sulfhydryl groups of some of the disulfide bonds of IgG results in the formation of two heavy, or H, chains (molecular weight 55,000) and two light, or L, chains (molecular weight 22,000). They are linked by disulfide bonds in the order L―H―H―L. Each H chain contains four intrachain disulfide bonds, and each L chain contains two. |
| |
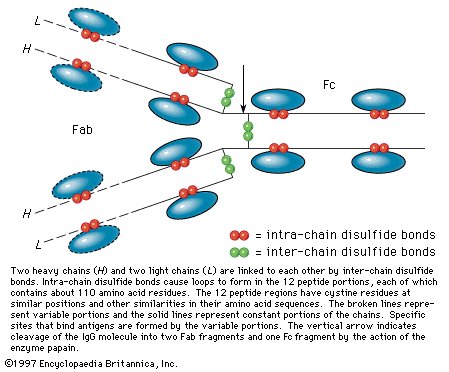 |
| |
| Figure 6: Diagram of an IgG immunoglobulin. |
| |
|
|
| |
|
|
Antibody preparations of the IgG type, even after removal of IgM and IgA antibodies, are heterogeneous. The H and L chains consist of a large number of different L chains and a variety of H chains. Pure IgG, IgM, and IgA immunoglobulins, however, occur in the blood serum of patients suffering from myelomas, which are malignant tumours of the bone marrow. The tumours produce either an IgG, an IgM, or an IgA protein, but rarely more than one class. A protein called the Bence-Jones protein, which is found in the urine of patients suffering from myeloma tumours, is identical with the L chains of the myeloma protein. Each patient has a different Bence-Jones protein; no two of the more than 100 Bence-Jones proteins that have been analyzed thus far are identical. It is thought that one lymphoid cell among hundreds of thousands becomes malignant and multiplies rapidly, forming the mass of a myeloma tumour that produces one γ-globulin.
Analyses of the Bence-Jones proteins have revealed that the L chains of humans and other mammals are of two quite different types, kappa (κ) and lambda (λ). Both consist of approximately 220 amino acids. The N–terminal halves of κ- and λ-chains are variable, differing in each Bence-Jones protein. The C–terminal halves of these same L chains have a constant amino acid sequence of either the κ- or the λ-type. The fact that one half of a peptide chain is variable and the other half invariant is contradictory to the view that the amino acid sequence of each peptide chain is determined by one gene. Evidently, two genes, one of them variable, the other invariant, fuse to form the gene for the single peptide chain of the L chains. Whereas the normal human L chains are always mixtures of the κ- and λ-types, the H chains of IgG, IgM, and IgA are different. They have been designated as gamma (γ), mu (μ), and alpha (α) chains, respectively. The N-terminal quarter of the H chains has a variable amino acid sequence; the C-terminal three-quarters of the H chains have a constant amino acid sequence.
Some of the amino acid sequences in the L and H chains are transmitted from generation to generation. As a result, the constant portion of the human L chains of the κ-type has in position 191 either valine or leucine. They correspond to two alleles (character-determining portions) of a gene; the two types are called allotypes. The valine-containing genetic type has been designated as InV(a+), the leucine-containing type as InV(b+). Many more allotypes, called Gm allotypes, have been found in the gamma chains of the human IgG immunoglobulins; more than 20 Gm allotypes are known. Certain combinations of Gm types occur. For example, the combination of Gm types 5, 6, and 11 has been found in Caucasians and African Americans but not in Chinese; the combination of 1, 2, and 17 has not been found in African Americans; and the combination of 1, 4, and 17 has not been found in Caucasians. Allotypes have also been discovered to occur in a number of other animals, including rabbits and mice.
It is understandable from the occurrence of a large number of allotypes that antibodies, even if produced in response to a single antigen, are mixtures of different allotypes. The existence of several classes of antibodies, of different allotypes, and of adaptation of the variable portions of antibodies to different regions of an antigen molecule results in a multiplicity of antibody molecules even if only a single antigen is administered. For this reason it has not yet been possible to unravel the amino acid sequence in the variable portion of antibody molecules. Much of the amino acid sequence in the constant regions of the L and H chains of humans and rabbit immunoglobulins, however, has been resolved. |

|
|
|
|
| |
Enzymes
|
Enzymes
Enzymes (B)
Practically all of the numerous and complex biochemical reactions that take place in animals, plants, and microorganisms are regulated by enzymes. These catalytic proteins are efficient and specific—that is, they accelerate the rate of one kind of chemical reaction of one type of compound, and they do so in a far more efficient manner than human-made catalysts. They are controlled by activators and inhibitors that initiate or block reactions. All cells contain enzymes, which usually vary in number and composition, depending on the cell type; an average mammalian cell, for example, is approximately one one-billionth (10−9) the size of a drop of water and generally contains about 3,000 enzymes.
The existence of enzymes was established in the middle of the 19th century by scientists studying the process of fermentation. The discovery of the role of enzymes as catalysts followed rapidly. Developments before 1850 included (in 1833) the separation from malt of the enzyme amylase, which converts starch into sugar, and (in 1836) the isolation from the stomach wall of animals of a component of gastric juice that could partially digest food in a test tube, the enzyme pepsin.
Enzymes were known for many years as ferments, a term derived from the Latin word for yeast. In 1878 the name enzyme, from the Greek words meaning “in yeast,” was introduced; since the late 19th century it has been employed universally. |

|
|
|
|
Role of enzymes in metabolism
Role of enzymes in metabolism (B)
Some enzymes help to break down large nutrient molecules, such as proteins, fats, and carbohydrates, into smaller molecules. This process occurs during the digestion of foodstuffs in the stomach and intestines of animals. Other enzymes guide the smaller, broken-down molecules through the intestinal wall into the bloodstream. Still other enzymes promote the formation of large, complex molecules from the small, simple ones to produce cellular constituents. Enzymes are also responsible for numerous other functions, which include the storage and release of energy, the course of reproduction, the processes of respiration, and vision. They are indispensable to life.
Each enzyme is able to promote only one type of chemical reaction. The compounds on which the enzyme acts are called substrates. Enzymes operate in tightly organized metabolic systems called pathways. A seemingly simple biological phenomenon—the contraction of a muscle, for example, or the transmission of a nerve impulse—actually involves a large number of chemical steps in which one or more chemical compounds (substrates) are converted to substances called products; the product of one step in a metabolic pathway serves as the substrate for the succeeding step in the pathway.
The role of enzymes in metabolic pathways can be illustrated diagrammatically. The chemical compound represented by A (see diagram below) is converted to product E in a series of enzyme-catalyzed steps, in which intermediate compounds represented by B, C, and D are formed in succession. They act as substrates for enzymes represented by 2, 3, and 4. Compound A may also be converted by another series of steps, some of which are the same as those in the pathway for the formation of E, to products represented by G and H.
The letters represent chemical compounds; numbers represent enzymes that catalyze individual reactions. The relative heights represent the thermodynamic energy of the compounds (e.g., compound A is more energy-rich than B, B more energy-rich than C). Compounds A, B, etc., change very slowly in the absence of a catalyst but do so rapidly in the presence of catalysts 1, 2, 3, etc.
The regulatory role of enzymes in metabolic pathways can be clarified by using a simple analogy: that between the compounds, represented by letters in the diagram, and a series of connected water reservoirs on a slope. Similarly, the enzymes represented by the numbers are analogous to the valves of the reservoir system. The valves control the flow of water in the reservoir; that is, if only valves 1, 2, 3, and 4 are open, the water in A flows only to E, but, if valves 1, 2, 5, and 6 are open, the water in A flows to G. In a similar manner, if enzymes 1, 2, 3, and 4 in the metabolic pathway are active, product E is formed, and, if enzymes 1, 2, 5, and 6 are active, product G is formed. The activity or lack of activity of the enzymes in the pathway therefore determines the fate of compound A; i.e., it either remains unchanged or is converted to one or more products. In addition, if products are formed, the activity of enzymes 3 and 4 relative to that of enzymes 5 and 6 determines the quantity of product E formed compared with product G.
Both the flow of water and the activity of enzymes obey the laws of thermodynamics; hence, water in reservoir F cannot flow freely to H by opening valve 7, because water cannot flow uphill. If, however, valves 1, 2, 5, and 7 are open, water flows from F to H, because the energy conserved during the downhill flow of water through valves 1, 2, and 5 is sufficient to allow it to force the water up through valve 7. In a similar way, enzymes in the metabolic pathway cannot convert compound F directly to H unless energy is available; enzymes are able to utilize energy from energy-conserving reactions in order to catalyze reactions that require energy. During the enzyme-catalyzed oxidation of carbohydrates to carbon dioxide and water, energy is conserved in the form of an energy-rich compound, adenosine triphosphate (ATP). The energy in ATP is utilized during an energy-consuming process such as the enzyme-catalyzed contraction of muscle.
Because the needs of cells and organisms vary, not only the activity but also the synthesis of enzymes must be regulated; e.g., the enzymes responsible for muscular activity in a leg muscle must be activated and inhibited at appropriate times. Some cells do not need certain enzymes; a liver cell, for example, does not need a muscle enzyme. A bacterium does not need enzymes to metabolize substances that are not present in its growth medium. Some enzymes, therefore, are not formed in certain cells, others are synthesized only when required, and still others are found in all cells. The formation and activity of enzymes are regulated not only by genetic mechanisms but also by organic secretions (hormones) from endocrine glands and by nerve impulses. Small molecules also play an important role (see below Enzyme flexibility and allosteric control).
If an enzyme is defective in some respect, disease may occur. The enzymes represented by the numbers 1 to 4 in the diagram must function during the conversion of the starting substance A to the product E. If one step is blocked because an enzyme is unable to function, product E may not be formed; if E is necessary for some vital function, disease results. Many inherited diseases and conditions of humans result from a deficiency of one enzyme. Some of these are listed in the table. Albinism, for example, results from an inherited lack of ability to synthesize the enzyme tyrosinase, which catalyzes one step in the pathway by which the pigment for hair and eye colour is formed. |
| |
Enzymes identified with hereditary diseases
| disease name |
defective enzyme |
| albinism |
tyrosinase |
| phenylketonuria |
phenylalanine hydroxylase |
| fructosuria |
fructokinase |
| methemoglobinemia |
methemoglobin reductase |
| galactosemia |
galactose-1-phosphate uridyl transferase |
|
|
| |
|
|

|
|
|
|
Other functions
Other functions (B)
Enzymes play an increasingly important role in medicine. The enzyme thrombin is used to promote the healing of wounds. Other enzymes are used to diagnose certain kinds of disease, to cause the remission of some forms of leukemia—a disease of the blood-forming organs—and to counteract unfavourable reactions in people who are allergic to penicillin. The enzyme lysozyme, which destroys cell walls, is used to kill bacteria. Enzymes have also been investigated for their potential to prevent tooth decay and to serve as anticoagulants in the treatment of thrombosis, a disease characterized by the formation of a clot, or plug, in a blood vessel. Enzymes may eventually be used to control enzyme deficiencies and abnormalities resulting from diseases.
It might also be noted in passing that enzymes are used in industrial processes involving the preparation of certain chemical compounds and the tanning of leather. They also are valuable in analytical procedures involving the detection of very small quantities of specific substances. Enzymes are necessary in various food-related industries, including cheese making, the brewing of beer, the aging of wine, and the baking of bread. Enzymes also may be used to clean clothes. For some industrial uses of enzymes, see baking. |

|
|
|
|
| |
General properties
|
Classification and nomenclature
Classification and nomenclature (B)
The first enzyme name, proposed in 1833, was diastase. Sixty-five years later, French microbiologist and chemist Émile Duclaux suggested that all enzymes be named by adding -ase to a root indicative of the nature of the substrate of the enzyme. Although enzymes are no longer named in such a simple manner, with the exception of a few—e.g., pepsin, trypsin, chymotrypsin, papain—most enzyme names do end in -ase.
Any systematic classification of enzymes should be based on a common property or quality that varies sufficiently to be useful as a distinguishing feature. In this regard, three properties of enzymes could serve as a basis for enzyme classification—the exact chemical nature of the enzyme, the chemical nature of the substrate, and the nature of the reaction catalyzed. In addition, although, as indicated above, early attempts at enzyme classification were based on the nature of broad groups of substrates (e.g., enzymes called carbohydrases act on carbohydrates), close functional similarities among enzymes in different groups were often obscured. By general agreement, enzymes now are classified according to their substrates and the nature of the reaction they catalyze.
In an attempt to devise a rational system of enzyme nomenclature, two names are given to an enzyme. One, known as the systematic name, is based on logical principles but is often long and awkward; the other, “trivial” name is short and generally used but not usually exact or systematic. In the scheme of systematic nomenclature, six main groups of enzymatic reactions are recognized; each catalyzes one reaction type and is subdivided on the basis of detailed definitions of the reaction catalyzed and of the substrate involved in the reaction. Enzymes that catalyze reactions in which hydrogen is transferred belong to the group known as oxidoreductases; those that catalyze the introduction of the elements of water at a specific site in a molecule are called hydrolases. The other four groups of reactions are the transferases—which catalyze reactions in which substances other than hydrogen are transferred—the lyases, the isomerases, and the ligases. Oxidoreductases and transferases account for about 50 percent of the approximately 1,000 enzymes recognized thus far. The table lists a few enzymes, their trivial names, their systematic names, and their biological roles. |
| |
Classification of some enzymes
| systematic name* |
trivial name |
reaction catalyzed |
biological role |
| code number** |
name*** |
|
|
|
| 1.1.1.1 |
alcohol: NAD oxidoreductase |
alcohol dehydrogenase |
alcohol + NAD → acetaldehyde NADH |
alcoholic fermentation |
| 1.1.1.27 |
L-lactate: NAD oxidoreductase |
lactic dehydrogenase |
lactate + NAD → pyruvate + NADH |
carbohydrate metabolism |
| 2.7.1.40 |
ATP: pyruvate phosphotransferase |
pyruvate kinase |
pyruvic acid + ATP → phosphoenolpyruvic acid + ADP |
carbohydrate metabolism |
| 3.1.1.7 |
acetylcholine: acetylhydrolase |
acetylcholinesterase |
acetylcholine + H2O → acetate + choline |
nerve-impulse conduction |
*Based on recommendations (1964) of the International Union of Biochemistry.
**The numbering system is as follows: the first number places the enzyme in one of six general groups—1, oxidoreductases; 2, transferases; 3, hydrolases; 4, lyases; 5, iomerases; and 6, ligases. The second number places the enzyme in a subclass based on substrate type or reaction type; e.g., the enzyme may act on molecules with −CHOH groups. The third number places the enzyme in a subsubclass, which specifies the reaction type more fully; e.g., NAD coenzyme required. The fourth number is the serial number of the enzyme in its subsubclass.
***NAD and NADH represent the oxidized and reduced forms of nicotinamide adenine dinucleotide (NAD), respectively; ATP and ADP represent adenosine triphosphate and adenosine diphosphate, respectively. |
|

|
|
|
|
Chemical nature
Chemical nature (B)
Little was known about the chemical nature of enzymes until the beginning of the 20th century, although scientists were almost convinced that they were proteins. In 1926 the enzyme urease was the first to be crystallized and clearly identified as a protein. Within the next few years the digestive enzymes pepsin, trypsin, and chymotrypsin were shown to be proteins. Since that time hundreds of enzymes, all of them proteins, have been prepared and characterized by chemical methods. Much of the knowledge of protein chemistry has, in fact, resulted from studies involving enzymes and from attempts to understand their nature and mode of action.
Although some enzymes consist of a single chain of the amino acids (i.e., simple organic molecules containing nitrogen), most enzymes are composed of more than one chain. Each chain is called a subunit. Many enzymes have two, four, or six subunits, and some consist of as many as 12 to 60 subunits. In many cases the subunits have identical structures; in others, however, several different types of subunit chains are involved.
With the exception of proteins that act as structural elements, most of the proteins in physiologically active tissues such as kidney and liver are enzymes. Regardless of the exact amount of enzymatic protein in an organism, it is clear that hundreds of different enzymes must be present in each tissue to account for the myriad reactions composing metabolism. |

|
|
|
|
Cofactors
Cofactors (B)
Although some enzymes consist only of protein, many are complex proteins; i.e., they have a protein component and a so-called cofactor. A complete enzyme is called a holoenzyme; if the cofactor is removed, the protein, no longer enzymatically active, is called the apoenzyme. A cofactor may be a metal—such as iron, copper, or magnesium—a moderately sized organic molecule called a prosthetic group, or a special type of substrate molecule known as a coenzyme. The cofactor may aid in the catalytic function of an enzyme, as do metals and prosthetic groups, or take part in the enzymatic reaction, as do coenzymes. |
| |
Functions of B-vitamin coenzymes in metabolism. |
|
|
| |
|
|
| A coenzyme serves as a type of substrate in certain enzymatic reactions and thus reacts in the exact proportions (i.e., stoichiometrically) required for reaction, rather than in catalytic quantities. A coenzyme may, for example, assume the role of a hydrogen acceptor, as does nicotinamide adenine dinucleotide (NAD), which accepts hydrogen from the substrate, or a chemical-group donor, as does adenosine triphosphate (ATP), which donates phosphoric acid to the substrate. After ATP has donated a phosphoric acid molecule to the substrate, the phosphoric acid can be reacquired in a second stoichiometric reaction catalyzed by a second enzyme. The catalytic nature of a coenzyme is apparent only when it couples the activities of two enzymes in this way. Coenzymes thus are the links, or shuttles, in metabolic pathways that enable substances—e.g., hydrogen, phosphoric acid—to be exchanged. |

|
|
|
|
| |
The nature of enzyme-catalyzed reactions
|
The nature of catalysis
The nature of catalysis (B)
In a chemical reaction—for example, one in which substance A is converted into product B—a point of equilibrium eventually is reached at which no further chemical change occurs; i.e., the rate of conversion of A to B equals the rate of conversion of B to A. The so-called thermodynamic-equilibrium constant expresses this chemical equilibrium. A catalyst may be defined as a substance that accelerates a chemical reaction but is not consumed in the process. The amount of catalyst has no relationship to the quantity of substance altered; very small amounts of enzymes are very efficient catalysts. Because the presence of an enzyme accelerates the rate of conversion of a compound to a product, it accelerates the approach to equilibrium; it does not, however, influence the equilibrium point attained.
The molecules in the watery medium of the cell are in constant thermal motion but, because they are more or less stable compounds, they would react only occasionally to form products in the absence of enzymes. There exists an energy barrier to the reaction of a molecule. The energy required to overcome the barrier to reaction is called the energy of activation. A reaction proceeds to equilibrium only if the molecules have sufficient energy of activation to form an activated complex, from which products can be derived. Enzymes greatly increase the chances for reactions by their ability to make large numbers of specific molecules more reactive (i.e., unstable) by forming intermediate compounds with them. The unstable intermediates quickly break down to form stable products, and the enzymes, unchanged by the reaction, are able to catalyze the formation of additional products. |

|
|
|
|
The role of the active site
The role of the active site (B)
That the compound on which an enzyme acts (substrate) must combine in some way with it before catalysis can proceed is an old idea, now supported by much experimental evidence. The combination of substrate molecules with enzymes involves collisions between the two. Enzymes are large molecules, the molecular weights of which (based on the weight of a hydrogen atom as 1) range from several thousand to several million. The substrates on which enzymes act usually have molecular weights of several hundred. Because of the difference in size between the two, only a fraction of the enzyme is in contact with the substrate; the region of contact is called the active site. Usually, each subunit of an enzyme has one active site capable of binding substrate.
The characteristics of an enzyme derive from the sequence of amino acids, which determine the shape of the enzyme (i.e., the structure of the active site) and hence the specificity of the enzyme. The forces that attract the substrate to the surface of an enzyme may be of a physical or a chemical nature. Electrostatic bonds may occur between oppositely charged groups—the circles containing plus and minus signs on the enzyme are attracted to their opposites in the substrate molecule. Such electrostatic bonds can occur with groups that are completely positively or negatively charged (i.e., ionic groups) or with groups that are partially charged (i.e., dipoles). The attractive forces between substrate and enzyme may also involve so-called hydrophobic bonds, in which the oily, or hydrocarbon, portions of the enzyme (represented by H-labelled circles) and the substrate are forced together in the same way as oil droplets tend to coalesce in water. |
| |
Enzyme; active site
The role of the active site in the lock-and-key fit of a substrate (the key) to an enzyme (the lock). |
|
|
| |
|
|
| Modifications in the structure of the amino acids at or near the active site usually affect the enzyme’s activity, because these amino acids are intimately involved in the fit and attraction of the substrate to the enzyme surface. The characteristics of the amino acids near the active site determine whether or not a substrate molecule will fit into the site. A molecule that is too bulky in the wrong places cannot fit into the active site and thus cannot react with the enzyme. In a similar manner, a molecule lacking essential attractive forces or the appropriately charged regions might not be bound to the enzyme. On the other hand, a molecule with a bulky group at a position such that it does not interfere with the binding of the molecule to the enzyme or with the function of the active site is able to serve as a substrate for the enzyme. The idea of a fit between substrate and enzyme, called the “ key–lock” hypothesis, was proposed by German chemist Emil Fischer in 1899 and explains one of the most important features of enzymes, their specificity. In most of the enzymes studied thus far, a cleft, or indentation, into which the substrate fits is found at the active site. |

|
|
|
|
The specificity of enzymes
The specificity of enzymes (B)
Since the substrate must fit into the active site of the enzyme before catalysis can occur, only properly designed molecules can serve as substrates for a specific enzyme; in many cases, an enzyme will react with only one naturally occurring molecule. Two oxidoreductase enzymes will serve to illustrate the principle of enzyme specificity. One (alcohol dehydrogenase) acts on alcohol, the other (lactic dehydrogenase) on lactic acid; the activities of the two, even though both are oxidoreductase enzymes, are not interchangeable—i.e., alcohol dehydrogenase will not catalyze a reaction involving lactic acid or vice versa, because the structure of each substrate differs sufficiently to prevent its fitting into the active site of the alternative enzyme. Enzyme specificity is essential because it keeps separate the many pathways, involving hundreds of enzymes, that function during metabolism.
Not all enzymes are highly specific. Digestive enzymes such as pepsin and chymotrypsin, for example, are able to act on almost any protein, as they must if they are to act upon the varied types of proteins consumed as food. On the other hand, thrombin, which reacts only with the protein fibrinogen, is part of a very delicate blood-clotting mechanism and thus must act only on one compound in order to maintain the proper functioning of the system.
When enzymes were first studied, it was thought that most of them were “absolutely specific”—that they would react with only one compound. In most cases, however, a molecule other than the natural substrate can be synthesized in the laboratory; it is enough like the natural substrate to react with the enzyme. Use of these synthetic substrates has been valuable in understanding enzymatic action. It must be remembered, however, that, in the living cell, many enzymes are absolutely specific for the compounds found there.
All enzymes isolated thus far are specific for the type of chemical reaction they catalyze—i.e., oxidoreductases do not catalyze hydrolase reactions, and hydrolases do not catalyze reactions involving oxidation and reduction. An enzyme therefore catalyzes a specific chemical reaction but may be able to do so on several similar compounds. |

|
|
|
|
The mechanism of enzymatic action
The mechanism of enzymatic action (B)
An enzyme attracts substrates to its active site, catalyzes the chemical reaction by which products are formed, and then allows the products to dissociate (separate from the enzyme surface). The combination formed by an enzyme and its substrates is called the enzyme–substrate complex. When two substrates and one enzyme are involved, the complex is called a ternary complex; one substrate and one enzyme are called a binary complex. The substrates are attracted to the active site by electrostatic and hydrophobic forces, which are called noncovalent bonds because they are physical attractions and not chemical bonds. |
| |
Figure 8: Mechanisms of enzymatic action (see text). |
|
|
| |
|
|
As an example, assume two substrates (S1 and S2) bind to the active site of the enzyme during step 1 and react to form products (P1 and P2) during step 2. The products dissociate from the enzyme surface in step 3, releasing the enzyme. The enzyme, unchanged by the reaction, is able to react with additional substrate molecules in this manner many times per second to form products. The step in which the actual chemical transformation occurs is of great interest, and, although much is known about it, it is not yet fully understood. In general there are two types of enzymatic mechanisms, one in which a so-called covalent intermediate forms and one in which none forms.
In the mechanism by which a covalent intermediate—i.e., an intermediate with a chemical bond between substrate and enzyme—forms, one substrate, B―X, for example, reacts with the group N on the enzyme surface to form an enzyme-B intermediate compound. The intermediate compound then reacts with the second substrate, Y, to form the products B―Y and X.
Many enzymes catalyze reactions by this type of mechanism. Acetylcholinesterase is used as a specific example in the sequence described below. The two substrates (S1 and S2) for acetylcholinesterase are acetylcholine (i.e., B―X) and water (Y). After acetylcholine (B―X) binds to the enzyme surface, a chemical bond forms between the acetyl moiety (B) of acetylcholine and the group N (part of the amino acid serine) on the enzyme surface. The result of the formation of this bond, called an acyl–serine bond, is one product, choline (X), and the enzyme-B intermediate compound (an acetyl–enzyme complex). The water molecule (Y) then reacts with the acyl–serine bond to form the second product, acetic acid (B―Y), which dissociates from the enzyme. Acetylcholinesterase is regenerated and is again able to react with another molecule of acetylcholine. This kind of reaction, involving the formation of an intermediate compound on the enzyme surface, is generally called a double displacement reaction.
Sucrose phosphorylase acts in a similar way. The substrate for sucrose phosphorylase is sucrose, or glucosyl-fructose (B―X), and the group N on the enzyme surface is a chemical group called a carboxyl group (COOH). The enzyme-B intermediate, a glucosyl–carboxyl compound, reacts with phosphate (Y) to form glucosyl-phosphate (B―Y). The other product (X) is fructose.
In double displacement reactions, the covalent intermediate between enzyme and substrate apparently influences the reaction to proceed more rapidly. Because the enzyme is unaltered at the end of the reaction, it functions as a true catalyst, even though it is temporarily altered during the enzymatic process.
Although many enzymes form a covalent intermediate, the mechanism is not essential for catalysis. One substrate (Y) reacts directly with the second substrate (X―B), in a so-called single displacement reaction. The B moiety, which is transformed in the chemical reaction, is involved in only one reaction and does not form a bond with a group on the enzyme surface. The enzyme maltose phosphorylase, for example, directly affects the bonds of the substrates (B―X and X), which, in this case, are maltose (glucosylglucose) and phosphate, to form the products, glucose (X) and glucosylphosphate (B―Y).
Covalent intermediates between part of a substrate and an enzyme occur in many enzymatic reactions, and various amino acids—serine, cysteine, lysine, and glutamic acid—are involved. |

|
|
|
|
| |
The rate of enzymatic reactions
|
The Michaelis-Menten hypothesis
The Michaelis-Menten hypothesis (B)
If the velocity of an enzymatic reaction is represented graphically as a function of the substrate concentration (S), the curve obtained in most cases is a hyperbola. The mathematical expression of this curve, shown in the equation below, was developed in 1912–13 by German biochemists Leonor Michaelis and Maud Leonora Menten. In the equation, VM is the maximal velocity of the reaction, and KM is called the Michaelis constant, |
| |
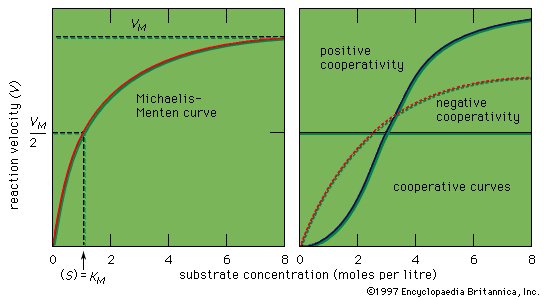
Figure 9: Curves representing enzyme action (see text). |
|
|
| |
|
|
|
The shape of the curve is a logical consequence of the active-site concept; i.e., the curve flattens at the maximum velocity (VM), which occurs when all the active sites of the enzyme are filled with substrate. The fact that the velocity approaches a maximum at high substrate concentrations provides support for the assumption that an intermediate enzyme–substrate complex forms. At the point of half the maximum velocity, the substrate concentration in moles per litre (M) is equal to the Michaelis constant, which is a rough measure of the affinity of the substrate molecule for the surface of the enzyme. KM values usually vary from about 10−8 to 10−2 M, and VM from 105 to 109 molecules of product formed per molecule of enzyme per second. The value for VM is referred to as the turnover number when expressed as moles of product formed per mole of enzyme per minute. The binding of molecules that inhibit or activate the protein surface usually results in similar types.
Enzymes are more efficient than human-made catalysts operating under the same conditions. Because many enzymes with different specificities occur in a cell, adequate space exists only for a few enzyme molecules catalyzing one specific reaction. Each enzyme, therefore, must be very efficient. One molecule of the enzyme catalase, for example, can produce 1012 molecules of oxygen per second. The catalytic groups at the active site of an enzyme act 106 to 109 times more effectively than do analogous groups in a nonenzymatic reaction.
The reason for the great efficiency of enzymes is not completely understood. It results in part from the precise positioning of the substrates and the catalytic groups at the active site, which serves to increase the probability of collision between the reacting atoms. In addition, the environment at the active site may be favourable for reaction—that is, acidic and basic groups may act together more effectively there, or some strain may be induced in the substrate molecules so that their bonds are broken more easily, or the orientation of the reacting substrates may be optimal at the enzyme surface. The theories that have been formulated to account for the high catalytic efficiency of enzymes, although reasonable, still remain to be proved. |

|
|
|
|
Inhibition of enzymes
Inhibition of enzymes (B)
Some molecules very similar to the substrate for an enzyme may be bound to the active site but be unable to react. Such molecules cover the active site and thus prevent the binding of the actual substrate to the site. This inhibition of enzyme action is of a competitive nature, because the inhibitor molecule actually competes with the substrate for the active site. The inhibitor sulfanilamide, for example, is similar enough to a substrate (p-aminobenzoic acid) of an enzyme involved in the metabolism of folic acid that it binds to the enzyme but cannot react. It covers the active site and prevents the binding of p-aminobenzoic acid. This enzyme is essential in certain disease-causing bacteria but is not essential to humans; large amounts of sulfanilamide therefore kill the microorganism but do not harm humans. Inhibitors such as sulfanilamide are called antimetabolites. Sulfanilamide and similar compounds that kill a pathogen without harming its host are widely used in chemotherapy. |
| |
|
Some inhibitors prevent, or block, enzymatic action by reacting with groups at the active site. The nerve gas diisopropyl fluorophosphate, for example, reacts with the serine at the active site of acetylcholinesterase to form a covalent bond. The nerve gas molecule involved in bond formation prevents the active site from binding the substrate, acetylcholine, thereby blocking catalysis and nerve action. Iodoacetic acid similarly blocks a key enzyme in muscle action by forming a bulky group on the amino acid cysteine, which is found at the enzyme’s active site. This process is called irreversible inhibition.
Some inhibitors modify amino acids other than those at the active site, resulting in loss of enzymatic activity. The inhibitor causes changes in the shape of the active site. Some amino acids other than those at the active site, however, can be modified without affecting the structure of the active site; in these cases, enzymatic action is not affected.
Such chemical changes parallel natural mutations. Inherited diseases frequently result from a change in an amino acid at the active site of an enzyme, thus making the enzyme defective. In some cases, an amino acid change alters the shape of the active site to the extent that it can no longer react; such diseases are usually fatal. In others, however, a partially defective enzyme is formed, and an individual may be very sick but able to live. |

|
|
|
|
Effects of temperature
Effects of temperature (B)
Enzymes function most efficiently within a physiological temperature range. Since enzymes are protein molecules, they can be destroyed by high temperatures. An example of such destruction, called protein denaturation, is the curdling of milk when it is boiled. Increasing temperature has two effects on an enzyme: first, the velocity of the reaction increases somewhat, because the rate of chemical reactions tends to increase with temperature; and, second, the enzyme is increasingly denatured. Increasing temperature thus increases the metabolic rate only within a limited range. If the temperature becomes too high, enzyme denaturation destroys life. Low temperatures also change the shapes of enzymes. With enzymes that are cold-sensitive, the change causes loss of activity. Both excessive cold and heat are therefore damaging to enzymes.
The degree of acidity or basicity of a solution, which is expressed as pH, also affects enzymes. As the acidity of a solution changes—i.e., the pH is altered—a point of optimum acidity occurs, at which the enzyme acts most efficiently. Although this pH optimum varies with temperature and is influenced by other constituents of the solution containing the enzyme, it is a characteristic property of enzymes. Because enzymes are sensitive to changes in acidity, most living systems are highly buffered; i.e., they have mechanisms that enable them to maintain a constant acidity. This acidity level, or pH, is about 7 in most organisms. Some bacteria function under moderately acidic or basic conditions; and the digestive enzyme pepsin acts in the acid milieu of the stomach. |

|
|
|
|
| |
Enzyme flexibility and allosteric control
|
The induced-fit theory
The induced-fit theory (B)
The key–lock hypothesis (see above The nature of enzyme-catalyzed reactions) does not fully account for enzymatic action; i.e., certain properties of enzymes cannot be accounted for by the simple relationship between enzyme and substrate proposed by the key–lock hypothesis. A theory called the induced-fit theory retains the key–lock idea of a fit of the substrate at the active site but postulates in addition that the substrate must do more than simply fit into the already preformed shape of an active site. Rather, the theory states, the binding of the substrate to the enzyme must cause a change in the shape of the enzyme that results in the proper alignment of the catalytic groups on its surface. This concept has been likened to the fit of a hand in a glove, the hand (substrate) inducing a change in the shape of the glove (enzyme). Although some enzymes appear to function according to the older key–lock hypothesis, most apparently function according to the induced-fit theory.
Typically, the substrate approaches the enzyme surface and induces a change in its shape that results in the correct alignment of the catalytic groups. In the case of the digestive enzyme carboxypeptidase, for example, the binding of the substrate causes a tyrosine molecule at the active site to move by as much as 15 angstroms. The catalytic groups at the active site react with the substrate to form products. The products separate from the enzyme surface, and the enzyme is able to repeat the sequence. Nonsubstrate molecules that are too bulky or too small alter the shape of the enzyme so that a misalignment of catalytic groups occurs; such molecules are not able to react even if they are attracted to the active site. |
| |
Figure 10: Induced-fit binding of a substrate to an enzyme surface and allosteric effects (see text). |
|
|
| |
|
|
The induced-fit theory explains a number of anomalous properties of enzymes. An example is “noncompetitive inhibition,” in which a compound inhibits the reaction of an enzyme but does not prevent the binding of the substrate. In this case, the inhibitor compound attracts the binding group so that the catalytic group is too far away from the substrate to react. The site at which the inhibitor binds to the enzyme is not the active site and is called an allosteric site. The inhibitor changes the shape of the active site to prevent catalysis without preventing binding of the substrate.
An inhibitor also can distort the active site by affecting the essential binding group; as a result, the enzyme can no longer attract the substrate. A so-called activator molecule affects the active site so that a nonsubstrate molecule is properly aligned and hence can react with the enzyme. Such activators can affect both binding and catalytic groups at the active site.
Enzyme flexibility is extremely important because it provides a mechanism for regulating enzymatic activity. The orientation at the active site can be disrupted by the binding of an inhibitor at a site other than the active site. Moreover, the enzyme can be activated by molecules that induce a proper alignment of the active site for a substrate that alone cannot induce this alignment.
As mentioned above, the sites that bind inhibitors and activators are called allosteric sites to distinguish them from active sites. Allosteric sites are in fact regulatory sites able to activate or inhibit enzymatic activity by influencing the shape of the enzyme. When the activator or inhibitor dissociates from the enzyme, it returns to its normal shape. Thus, the flexibility of the protein structure allows the operation of a simple, reversible control system similar to a thermostat. |

|
|
|
|
Types of allosteric control
Types of allosteric control (B)
Allosteric control can operate in many ways; two examples serve to illustrate some general effects. A pathway consisting of ten enzymes is involved in the synthesis of the amino acid histidine. When a cell contains enough histidine, synthesis stops—an appropriate economy move by the cell. Synthesis is stopped by the inhibition of the first enzyme in the pathway by the product, histidine. The inhibition of an enzyme by a product is called feedback inhibition; i.e., a product many steps removed from an initial enzyme blocks its action. Feedback inhibition occurs in many pathways in all living things.
Allosteric control can also be achieved by activators. The hormone adrenaline (epinephrine) acts in this way. When energy is needed, adrenaline is released and activates, by allosteric activation, the enzyme adenyl cyclase. This enzyme catalyzes a reaction in which the compound cyclic adenosine monophosphate (cyclic AMP) is formed from ATP. Cyclic AMP in turn acts as an allosteric activator of enzymes that speed the metabolism of carbohydrate to produce energy. This type of allosteric regulation also is widespread in biological systems. Thus, a combination of allosteric activation and inhibition allows the production of energy or materials when they are needed and shuts off production when the supply is adequate.
Allosteric control is a rapid method of regulating products continuously needed by living things. Yet some cells have no need for certain enzymes, and it would be wasteful for the cell to synthesize them. In this case, certain molecules, called repressors, prevent the synthesis of unneeded enzymes. The repressors are proteins that bind to DNA and prevent the first step in the process resulting in protein synthesis. If certain metabolites are added to cells that need an enzyme, enzyme synthesis occurs—i.e., it is induced. Addition of galactose to a growth medium containing Escherichia coli bacteria, for example, induces the synthesis of the enzyme beta-galactosidase. The bacteria thus can synthesize this galactose-metabolizing enzyme when it is needed and prevent its synthesis when it is not. The way in which the synthesis of enzymes is induced or repressed in mammalian systems is less understood but is believed to be similar.
Different types of cells in complex organisms have different enzymes, even though they have the same DNA content. The enzymes actually synthesized are the ones needed in a specific cell and vary not only for different types of cells—e.g., nerve, muscle, eye, and skin cells—but also for different species.
In an enzyme consisting of several subunits, or chains, alteration in the shape of one chain as a result of the influence either of a substrate molecule or of allosteric inhibitors or activators may change the shape of a neighbouring chain. As a result, the binding of a second molecule of substrate occurs in a different way from the binding of the first, and the third is different from the second. This phenomenon, called cooperativity, is characteristic of allosteric enzymes. Cooperativity is reflected by a sigmoid curve, as compared to the hyperbolic curve of Michaelis–Menten. An enzyme of several subunits that exhibits cooperativity is far more sensitive to control mechanisms than is an enzyme of one subunit and hence one active site.
The first example of cooperativity was observed in hemoglobin, which is not an enzyme but behaves like one in many ways. The absorption of oxygen in the lungs and its deposition in the tissues is far more efficient because the subunits of hemoglobin show positive cooperativity, so called because the first molecule of substrate makes it easier for the next to bind.
Negative cooperativity, in which the binding of one molecule makes it less easy for the next to bind, also occurs in living things. Negative cooperativity makes an enzyme less sensitive to fluctuations in concentrations of metabolites and may be important for enzymes that must be present in the cell at relatively constant levels of activity.
Some enzymes are closely associated aggregates of several enzyme units; the pyruvate dehydrogenase system, for example, contains five different enzymes, has a total molecular weight of 4,000,000, and consists of four different types of chains. Apparently, the enzymes in cells may be organized by forming complex units, by being absorbed on a cell wall, or by being isolated by membranes in special compartments. Since a pathway involves the stepwise modification of chemical compounds, aggregations of the enzymes in a given pathway facilitate their function in a manner similar to an industrial assembly line. |

|
|
|
|

|
|
|
|
|
|

